Bioactive Metabolites from Terrestrial and Marine Actinomycetes
Abstract
:1. Introduction
2. Ecology of Actinomycetes
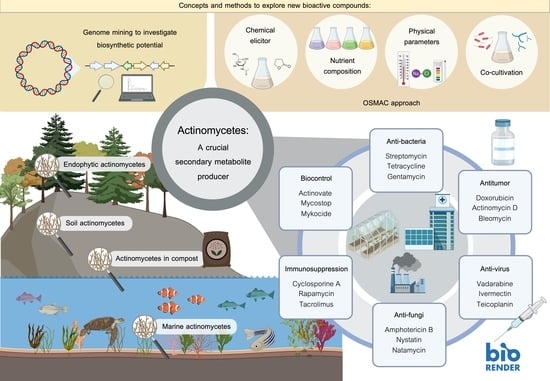
2.1. Soil Actinomycetes
2.2. Endophytic Actinomycetes
2.3. Actinomycetes in Compost
2.4. Marine Actinomycetes
3. Taxonomy and Classification
4. A Crucial Secondary Metabolite Producer
5. Biological Activity of Secondary Metabolites from Actinomycetes and Their Applications
5.1. Antibacterial Agents
5.2. Antifungal Agents
5.3. Immunosuppressive Agent
5.4. Biocontrol Agents
| Commercial Product Name | Organism As Active Substance | Registered As a Microbial Pesticide | Targeted Pest/Pathogen/Disease |
|---|---|---|---|
| Actinovate, Novozymes BioAg Inc., Milwaukee, WI, USA | S. lydicus WYEC 108 | Canada, USA | Soilborne diseases, viz. Pythium, Fusarium, Phytophthora, Rhizoctonia, and Verticillium; foliar diseases such as powdery and downy mildew, Botrytis, Alternaria, Postia, Geotrichum, and Sclerotinia |
| Mycostop, Verdera Oy, Espoo, Finland | Streptomyces K61 | EU, Canada, USA | Damping off caused by Alternaria, R. solani, Fusarium, Phytophthora, Pythium wilt, and root diseases |
| Mykocide, KIBC Co., Ltd., Yongin, Gyeonggi-do, Republic of Korea | S. colombiensis | Republic of Korea | Powdery mildews, grey mold, and brown patch |
| Bactophil | Streptomyces albus | Ukraine | Seed germination diseases |
5.5. Antitumor Compounds
5.6. Antiviral Agents
5.7. Other Activities
6. Concepts and Methods to Explore New Bioactive Compounds
6.1. Exploring New Habitats or Extreme Environments As A Source for Novel Strains
6.2. Genome Mining to Investigate Biosynthetic Potential
6.3. OSMAC Approach
6.4. Co-Cultivation Technique
6.5. Using Chemical Elicitors
6.5.1. γ-. Butyrolactones and Related Regulators
6.5.2. N-Acetylglucosamine (GlcNAc)
7. Future Perspectives on Actinomycetes
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kieser, T.; Bibb, M.J.; Buttner, M.J.; Chater, K.F.; Hopwood, D.A. Practical Streptomyces Genetics; The John Innes Foundation: Norwich, UK, 2000. [Google Scholar]
- Tiwari, K.; Gupta, R.K. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 2012, 32, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Ding, T.; Yang, L.-J.; Zhang, W.-D.; Shen, Y.-H. The secondary metabolites of rare actinomycetes: Chemistry and bioactivity. RSC Adv. 2019, 9, 21964–21988. [Google Scholar] [CrossRef] [PubMed]
- Al-Fadhli, A.A.; Threadgill, M.D.; Mohammed, F.; Sibley, P.; Al-Ariqi, W.; Parveen, I. Macrolides from rare actinomycetes: Structures and bioactivities. Int. J. Antimicrob. Agents 2022, 59, 106523. [Google Scholar] [CrossRef] [PubMed]
- Prudence, S.M.M.; Addington, E.; Castaño-Espriu, L.; Mark, D.R.; Pintor-Escobar, L.; Russell, A.H.; McLean, T.C. Advances in actinomycete research: An ActinoBase review of 2019. Microbiology 2020, 166, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Qinyuan, L.; Xiu, C.; Yi, J.; Chenglin, J. Morphological Identification of Actinobacteria. In Actinobacteria; Dharumadurai, D., Yi, J., Eds.; IntechOpen: Rijeka, Croatia, 2016; pp. 59–86. [Google Scholar]
- Goodfellow, M.; Williams, S.T. Ecology of actinomycetes. Annu. Rev. Microbiol. 1983, 37, 189–216. [Google Scholar] [CrossRef]
- van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, A.; Takahashi, Y. Endophytic actinomycetes: Promising source of novel bioactive compounds. J. Ant. 2017, 70, 514–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helmke, E.; Weyland, H. Rhodococcus marinonascens sp. nov., an actinomycete from the sea. Int. J. Syst. Bacteriol. 1984, 34, 127–138. [Google Scholar] [CrossRef] [Green Version]
- Masand, M.; Jose, P.A.; Menghani, E.; Jebakumar, S.R.D. Continuing hunt for endophytic actinomycetes as a source of novel biologically active metabolites. World J. Microbiol. 2015, 31, 1863–1875. [Google Scholar] [CrossRef]
- Kumar, S.; Solanki, D.S.; Parihar, K.; Tak, A.; Gehlot, P.; Pathak, R.; Singh, S.K. Actinomycetes isolates of arid zone of Indian Thar Desert and efficacy of their bioactive compounds against human pathogenic bacteria. Biol. Futur. 2021, 72, 431–440. [Google Scholar] [CrossRef]
- Mohammadipanah, F.; Wink, J. Actinobacteria from arid and desert habitats: Diversity and biological activity. Front. Microbiol. 2016, 6, 1541. [Google Scholar] [CrossRef] [Green Version]
- Silva, L.J.; Crevelin, E.J.; Souza, D.T.; Lacerda-Júnior, G.V.; de Oliveira, V.M.; Ruiz, A.L.T.G.; Rosa, L.H.; Moraes, L.A.B.; Melo, I.S. Actinobacteria from Antarctica as a source for anticancer discovery. Sci. Rep. 2020, 10, 13870. [Google Scholar] [CrossRef]
- Zenova, G.M.; Manucharova, N.A.; Zvyagintsev, D.G. Extremophilic and extremotolerant actinomycetes in different soil types. Eurasian Soil Sci. 2011, 44, 417–436. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Aamir, M.; Rai, K.K.; Zehra, A.; Dubey, M.K.; Samal, S.; Yadav, M.; Upadhyay, R.S. Endophytic actinomycetes in bioactive compounds production and plant defense system. In Microbial Endophytes; Kumar, A., Singh, V.K., Eds.; Woodhead Publishing: Sawston, UK, 2020; pp. 189–229. [Google Scholar] [CrossRef]
- Chaurasia, A.; Meena, B.R.; Tripathi, A.N.; Pandey, K.K.; Rai, A.B.; Singh, B. Actinomycetes: An unexplored microorganisms for plant growth promotion and biocontrol in vegetable crops. World J. Microbiol. 2018, 34, 132. [Google Scholar] [CrossRef] [PubMed]
- Nonthakaew, N.; Panbangred, W.; Songnuan, W.; Intra, B. Plant growth-promoting properties of Streptomyces spp. isolates and their impact on mung bean plantlets’ rhizosphere microbiome. Front. Microbiol 2022, 13, 967415. [Google Scholar] [CrossRef] [PubMed]
- Intra, B.; Mungsuntisuk, I.; Nihira, T.; Igarashi, Y.; Panbangred, W. Identification of actinomycetes from plant rhizospheric soils with inhibitory activity against Colletotrichum spp., the causative agent of anthracnose disease. BMC Res. Notes 2011, 4, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, Y.; Dolfing, J.; Guo, Z.; Chen, R.; Wu, M.; Li, Z.; Lin, X.; Feng, Y. Important ecophysiological roles of non-dominant Actinobacteria in plant residue decomposition, especially in less fertile soils. Microbiome 2021, 9, 84. [Google Scholar] [CrossRef]
- Guan, T.W.; Lin, Y.J.; Ou, M.Y.; Chen, K.B. Isolation and diversity of sediment bacteria in the hypersaline aiding lake, China. PLoS ONE 2020, 15, e0236006. [Google Scholar] [CrossRef]
- Ahmed, V.; Verma, M.K.; Gupta, S.; Mandhan, V.; Chauhan, N.S. Metagenomic Profiling of Soil Microbes to Mine Salt Stress Tolerance Genes. Front. Microb. 2018, 9, 159. [Google Scholar] [CrossRef] [Green Version]
- Rungin, S.; Indananda, C.; Suttiviriya, P.; Kruasuwan, W.; Jaemsaeng, R.; Thamchaipenet, A. Plant growth enhancing effects by a siderophore-producing endophytic streptomycete isolated from a Thai jasmine rice plant (Oryza sativa L. cv. KDML105). Antonie Van Leeuwenhoek 2012, 102, 463–472. [Google Scholar] [CrossRef]
- Janso, J.E.; Carter, G.T. Biosynthetic Potential of Phylogenetically Unique Endophytic Actinomycetes from Tropical Plants. Appl. Environ. Microbiol. 2010, 76, 4377–4386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayakawa, M. Studies on the isolation and distribution of rare actinomycetes in soil. Actinomycetologica 2008, 22, 12–19. [Google Scholar] [CrossRef] [Green Version]
- Hemati, A.; Nazari, M.; Asgari Lajayer, B.; Smith, D.L.; Astatkie, T. Lignocellulosics in plant cell wall and their potential biological degradation. Folia Microbiol. 2022, 67, 671–681. [Google Scholar] [CrossRef]
- Hemati, A.; Aliasgharzad, N.; Khakvar, R.; Delangiz, N.; Asgari Lajayer, B.; van Hullebusch, E.D. Bioaugmentation of thermophilic lignocellulose degrading bacteria accelerate the composting process of lignocellulosic materials. Biomass Conv. Bioref. 2022, 12, 1–15. [Google Scholar] [CrossRef]
- Hemati, A.; Aliasgharzad, N.; Khakvar, R.; Khoshmanzar, E.; Asgari Lajayer, B.; van Hullebusch, E.D. Role of lignin and thermophilic lignocellulolytic bacteria in the evolution of humification indices and enzymatic activities during compost production. Waste Manag. 2021, 119, 122–134. [Google Scholar] [CrossRef] [PubMed]
- de Gannes, V.; Eudoxie, G.; Hickey, W.J. Prokaryotic successions and diversity in composts as revealed by 454-pyrosequencing. Bioresour. Technol. 2013, 133, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Takaku, H.; Kodaira, S.; Kimoto, A.; Nashimoto, M.; Takagi, M. Microbial communities in the garbage composting with rice hull as an amendment revealed by culture-dependent and -independent approaches. J. Biosci. Bioeng. 2006, 101, 42–50. [Google Scholar] [CrossRef]
- Setyati, W.A.; Pringgenies, D.; Soenardjo, N.; Pramesti, R. Actinomycetes of secondary metabolite producers from mangrove sediments, Central Java, Indonesia. Vet. World 2021, 14, 2620. [Google Scholar] [CrossRef]
- Khalifa, S.A.M.; Elias, N.; Farag, M.A.; Chen, L.; Saeed, A.; Hegazy, M.E.F.; Moustafa, M.S.; El-Wahed, A.A.; Al-Mousawi, S.M.; Musharraf, S.G.; et al. Marine Natural Products: A Source of Novel Anticancer Drugs. Mar. Drugs 2019, 17, 491. [Google Scholar] [CrossRef] [Green Version]
- Jensen, P.R.; Dwight, R.; Fenical, W. Distribution of actinomycetes in near-shore tropical marine sediments. Appl. Environ. Microbiol. 1991, 57, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Jensen, P.R.; Mafnas, C. Biogeography of the marine actinomycete Salinispora. Environ. Microbiol. 2006, 8, 1881–1888. [Google Scholar] [CrossRef]
- Bienhold, C.; Zinger, L.; Boetius, A.; Ramette, A. Diversity and Biogeography of Bathyal and Abyssal Seafloor Bacteria. PLoS ONE 2016, 11, e0148016. [Google Scholar] [CrossRef] [PubMed]
- Betancur, L.A.; Naranjo-Gaybor, S.J.; Vinchira-Villarraga, D.M.; Moreno-Sarmiento, N.C.; Maldonado, L.A.; Suarez-Moreno, Z.R.; Acosta-González, A.; Padilla-Gonzalez, G.F.; Puyana, M.; Castellanos, L.; et al. Marine Actinobacteria as a source of compounds for phytopathogen control: An integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS ONE 2017, 12, e0170148. [Google Scholar] [CrossRef] [Green Version]
- Mahmoud, H.M.; Kalendar, A.A. Coral-associated Actinobacteria: Diversity, abundance, and biotechnological potentials. Front. Microb. 2016, 7, 204. [Google Scholar] [CrossRef] [Green Version]
- Sarmiento-Vizcaíno, A.; González, V.; Braña, A.F.; Palacios, J.J.; Otero, L.; Fernández, J.; Molina, A.; Kulik, A.; Vázquez, F.; Acuña, J.L.; et al. Pharmacological Potential of Phylogenetically Diverse Actinobacteria Isolated from Deep-Sea Coral Ecosystems of the Submarine Avilés Canyon in the Cantabrian Sea. Microb. Ecol. 2017, 73, 338–352. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ye, Y.; Wang, R.; Zhang, Y.; Wu, C.; Debnath, S.C.; Ma, Z.; Wang, J.; Wu, M. Streptomyces nigra sp. nov. Is a Novel Actinobacterium Isolated From Mangrove Soil and Exerts a Potent Antitumor Activity in Vitro. Front. Microbiol. 2018, 9, 1587. [Google Scholar] [CrossRef] [Green Version]
- Kemung, H.M.; Tan, L.T.H.; Chan, K.G.; Ser, H.L.; Law, J.W.F.; Lee, L.H.; Goh, B.H. Streptomyces sp. Strain MUSC 125 from Mangrove Soil in Malaysia with Anti-MRSA, Anti-Biofilm and Antioxidant Activities. Molecules 2020, 25, 3545. [Google Scholar] [CrossRef]
- Lin, X.; Hetharua, B.; Lin, L.; Xu, H.; Zheng, T.; He, Z.; Tian, Y. Mangrove Sediment Microbiome: Adaptive Microbial Assemblages and Their Routed Biogeochemical Processes in Yunxiao Mangrove National Nature Reserve, China. Microb. Ecol. 2019, 78, 57–69. [Google Scholar] [CrossRef]
- Xu, D.; Han, L.; Li, C.; Cao, Q.; Zhu, D.; Barrett, N.H.; Harmody, D.; Chen, J.; Zhu, H.; McCarthy, P.J.; et al. Bioprospecting deep-sea actinobacteria for novel anti-infective natural products. Front. Microbiol. 2018, 9, 787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olano, C.; Méndez, C.; Salas, J.A. Antitumor compounds from marine actinomycetes. Mar. Drugs 2009, 7, 210–248. [Google Scholar] [CrossRef] [Green Version]
- Subramani, R.; Aalbersberg, W. Marine actinomycetes: An ongoing source of novel bioactive metabolites. Microbiol. Res. 2012, 167, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Anderson, A.S.; Wellington, E.M.H. The taxonomy of Streptomyces and related genera. Int. J. Syst. Evol. Microbiol. 2001, 51, 797–814. [Google Scholar] [CrossRef] [Green Version]
- Ngamcharungchit, C.; Kanto, H.; Také, A.; Intra, B.; Matsumoto, A.; Panbangred, W.; Inahashi, Y. Amycolatopsis iheyensis sp. nov., isolated from soil on Iheya Island, Japan. Int. J. Syst. Evol. Microbiol. 2023, 73, 005757. [Google Scholar] [CrossRef] [PubMed]
- Citarella, R.V.; Colwell, R.R. Polyphasic Taxonomy of the Genus Vibrio: Polynucleotide Sequence Relationships Among Selected Vibrio Species. J. Bacteriol. 1970, 104, 434. [Google Scholar] [CrossRef]
- Vandamme, P.; Pot, B.; Gillis, M.; De Vos, P.; Kersters, K.; Swings, J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 1996, 60, 407–438. [Google Scholar] [CrossRef]
- Wayne, L.G.; Brenner, D.J.; Colwell, R.R.; Grimont, P.A.D.; Kandler, O.; Krichevsky, M.I.; Moore, L.H.; Moore, W.E.C.; Murray, R.G.E.; Stackebrandt, E.; et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int. J. Syst. Evol. Microbiol. 1987, 37, 463–464. [Google Scholar] [CrossRef] [Green Version]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. Microbiol. 2010, 60, 249–266. [Google Scholar] [CrossRef] [Green Version]
- Intra, B.; Matsumoto, A.; Inahashi, Y.; Ōmura, S.; Takahashi, Y.; Panbangred, W. Actinokineospora bangkokensis sp. nov., isolated from rhizospheric soil. Int. J. Syst. Evol. Microbiol. 2013, 63, 2655–2660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Intra, B.; Euanorasetr, J.; Také, A.; Inahashi, Y.; Mori, M.; Panbangred, W.; Matsumoto, A. Saccharopolyspora rhizosphaerae sp. nov., an actinomycete isolated from rhizosphere soil in Thailand. Int. J. Syst. Evol. Microbiol. 2019, 69, 1299–1305. [Google Scholar] [CrossRef] [PubMed]
- Intra, B.; Matsumoto, A.; Inahashi, Y.; Ōmura, S.; Panbangred, W.; Takahashi, Y. Streptosporangium jomthongense sp. nov., an actinomycete isolated from rhizospheric soil and emendation of the genus Streptosporangium. Int. J. Syst. Evol. Microbiol. 2014, 64, 2400–2406. [Google Scholar] [CrossRef] [Green Version]
- Wattanasuepsin, W.; Intra, B.; Také, A.; Inahashi, Y.; Euanorasetr, J.; Ōmura, S.; Matsumoto, A.; Panbangred, W. Saccharomonospora colocasiae sp. Nov., an actinomycete isolated from the rhizosphere of Colocasia esculenta. Int. J. Syst. Evol. Microbiol. 2017, 67, 4572–4577. [Google Scholar] [CrossRef]
- Intra, B.; Panbangred, W.; Inahashi, Y.; Také, A.; Mori, M.; Ōmura, S.; Matsumoto, A. Micromonospora pelagivivens sp. nov., a new species of the genus Micromonospora isolated from deep-sea sediment in Japan. Int. J. Syst. Evol. Microbiol. 2020, 70, 3069–3075. [Google Scholar] [CrossRef]
- Bérdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading? J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [Green Version]
- Challis, G.L.; Hopwood, D.A. Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc. Nat. Acad. Sci. USA 2003, 100, 14555–14561. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; De Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Cdc. Antibiotic Resistance Threats Report In The United States 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 16 June 2023).
- De Simeis, D.; Serra, S. Actinomycetes: A Never-Ending Source of Bioactive Compounds—An Overview on Antibiotics Production. Antibiotics 2021, 10, 483. [Google Scholar] [CrossRef]
- Euanorasetr, J.; Nilvongse, A.; Tantimavanich, S.; Nihira, T.; Igarashi, Y.; Panbangred, W. Identification and characterization of soil-isolated Streptomyces SJE177 producing actinomycin. Southeast Asian J. Trop. Med. Public Health 2010, 41, 1177–1187. [Google Scholar]
- Euanorasetr, J.; Intra, B.; Mongkol, P.; Chankhamhaengdecha, S.; Tuchinda, P.; Mori, M.; Shiomi, K.; Nihira, T.; Panbangred, W. Spirotetronate antibiotics with anti-Clostridium activity from Actinomadura sp. 2EPS. W. J. Microbiol. Biotech. 2015, 31, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Iorio, M.; Cruz, J.; Simone, M.; Bernasconi, A.; Brunati, C.; Sosio, M.; Donadio, S.; Maffioli, S.I. Antibacterial Paramagnetic Quinones from Actinoallomurus. J. Nat. Prod. 2017, 80, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Kodani, S.; Komaki, H.; Ishimura, S.; Hemmi, H.; Ohnishi-Kameyama, M. Isolation and structure determination of a new lantibiotic cinnamycin B from Actinomadura atramentaria based on genome mining. J. Ind. Microbiol. Biotechnol. 2016, 43, 1159–1165. [Google Scholar] [CrossRef] [Green Version]
- Shin, B.; Kim, B.Y.; Cho, E.; Oh, K.B.; Shin, J.; Goodfellow, M.; Oh, D.C. Actinomadurol, an Antibacterial Norditerpenoid from a Rare Actinomycete, Actinomadura sp. KC 191. J. Nat. Prod. 2016, 79, 1886–1890. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Calil, F.A.; Pinto, F.d.C.L.; Medeiros, T.C.T.; Almeida, L.C.; Silva, L.J.; de Melo, I.S.; Zucchi, T.D.; Costa-Lotufo, L.V.; Moraes, L.A.B. Pradimicin-IRD from Amycolatopsis sp. IRD-009 and its antimicrobial and cytotoxic activities. Nat. Prod. Res. 2019, 33, 1713–1720. [Google Scholar] [CrossRef] [PubMed]
- Beemelmanns, C.; Ramadhar, T.R.; Kim, K.H.; Klassen, J.L.; Cao, S.; Wyche, T.P.; Hou, Y.; Poulsen, M.; Bugni, T.S.; Currie, C.R.; et al. Macrotermycins A-D, Glycosylated Macrolactams from a Termite-Associated Amycolatopsis sp. M39. Org. Lett. 2017, 19, 1000–1003. [Google Scholar] [CrossRef]
- Khan, A.; Said, M.S.; Borade, B.R.; Gonnade, R.; Barvkar, V.; Kontham, R.; Dastager, S.G. Enceleamycins A-C, Furo-Naphthoquinones from Amycolatopsis sp. MCC0218: Isolation, Structure Elucidation, and Antimicrobial Activity. J. Nat. Prod. 2022, 85, 1267–1273. [Google Scholar] [CrossRef]
- Hashizume, H.; Sawa, R.; Yamashita, K.; Nishimura, Y.; Igarashi, M. Structure and antibacterial activities of new cyclic peptide antibiotics, pargamicins B, C and D, from Amycolatopsis sp. ML1-hF4. J. Ant. 2017, 70, 699–704. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Liang, X.; Zhang, S.; Song, Z.; Wang, C.; Xu, Y. Maipomycin A, a Novel Natural Compound With Promising Anti-biofilm Activity Against Gram-Negative Pathogenic Bacteria. Front. Microbiol. 2021, 11, 598024. [Google Scholar] [CrossRef]
- Kohda, Y.; Sakamoto, S.; Umekita, M.; Kimura, T.; Kubota, Y.; Arisaka, R.; Shibuya, Y.; Muramatsu, H.; Sawa, R.; Dan, S.; et al. Isolation of new derivatives of the 20-membered macrodiolide bispolide from Kitasatospora sp. MG372-hF19. J. Antibiot. 2021, 75, 77–85. [Google Scholar] [CrossRef]
- Uzair, B.; Menaa, F.; Khan, B.A.; Mohammad, F.V.; Ahmad, V.U.; Djeribi, R.; Menaa, B. Isolation, purification, structural elucidation and antimicrobial activities of kocumarin, a novel antibiotic isolated from actinobacterium Kocuria marina CMG S2 associated with the brown seaweed Pelvetia canaliculata. Microbiol. Res. 2018, 206, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Gong, T.; Zhen, X.; Li, X.L.; Chen, J.J.; Chen, T.J.; Yang, J.L.; Zhu, P. Tetrocarcin Q, a New Spirotetronate with a Unique Glycosyl Group from a Marine-Derived Actinomycete Micromonospora carbonacea LS276. Mar. Drugs 2018, 16, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, Y.H.; Bae, S.; Sim, J.; Hur, J.; Jo, S.I.; Shin, J.; Suh, Y.G.; Oh, K.B.; Oh, D.C. Nicrophorusamides A and B, Antibacterial Chlorinated Cyclic Peptides from a Gut Bacterium of the Carrion Beetle Nicrophorus concolor. J. Nat. Prod. 2017, 80, 2962–2968. [Google Scholar] [CrossRef] [PubMed]
- Gui, C.; Zhang, S.; Zhu, X.; Ding, W.; Huang, H.; Gu, Y.C.; Duan, Y.; Ju, J. Antimicrobial Spirotetronate Metabolites from Marine-Derived Micromonospora harpali SCSIO GJ089. J. Nat. Prod. 2017, 80, 1594–1603. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xie, L.; Zhao, W.; Zhou, J.; Jiang, H.; Liu, W.; Jiang, H.; Lin, F. Two new rakicidin derivatives from marine Micromonospora chalcea FIM-R160609. Nat. Prod. Res. 2022, 36, 1–8. [Google Scholar] [CrossRef]
- Hifnawy, M.S.; Hassan, H.M.; Mohammed, R.; Fouda, M.M.; Sayed, A.M.; Hamed, A.A.; AbouZid, S.F.; Rateb, M.E.; Alhadrami, H.A.; Abdelmohsen, U.R. Induction of Antibacterial Metabolites by Co-Cultivation of Two Red-Sea-Sponge-Associated Actinomycetes Micromonospora sp. UR56 and Actinokinespora sp. EG49. Mar. Drugs 2020, 18, 243. [Google Scholar] [CrossRef] [PubMed]
- Adnani, N.; Chevrette, M.G.; Adibhatla, S.N.; Zhang, F.; Yu, Q.; Braun, D.R.; Nelson, J.; Simpkins, S.W.; McDonald, B.R.; Myers, C.L.; et al. Coculture of Marine Invertebrate-Associated Bacteria and Interdisciplinary Technologies Enable Biosynthesis and Discovery of a New Antibiotic, Keyicin. ACS Chem. Biol. 2017, 12, 3093–3102. [Google Scholar] [CrossRef]
- Tan, Y.; Hu, Y.; Wang, Q.; Zhou, H.; Wang, Y.; Gan, M. Tetrocarcins N and O, glycosidic spirotetronates from a marine-derived Micromonospora sp. identified by PCR-based screening. RSC Adv. 2016, 6, 91773–91778. [Google Scholar] [CrossRef]
- Pérez-Bonilla, M.; Oves-Costales, D.; De La Cruz, M.; Kokkini, M.; Martín, J.; Vicente, F.; Genilloud, O.; Reyes, F. Phocoenamicins B and C, New Antibacterial Spirotetronates Isolated from a Marine Micromonospora sp. Mar. Drugs 2018, 16, 95. [Google Scholar] [CrossRef] [Green Version]
- Williams, D.E.; Dalisay, D.S.; Chen, J.; Polishchuck, E.A.; Patrick, B.O.; Narula, G.; Ko, M.; Av-Gay, Y.; Li, H.; Magarvey, N.; et al. Aminorifamycins and Sporalactams Produced in Culture by a Micromonospora sp. Isolated from a Northeastern-Pacific Marine Sediment Are Potent Antibiotics. Org. Lett. 2017, 19, 766–769. [Google Scholar] [CrossRef]
- Cheng, Z.; Zhang, Q.; Peng, J.; Zhao, X.; Ma, L.; Zhang, C.; Zhu, Y. Genomics-Driven Discovery of Benzoxazole Alkaloids from the Marine-Derived Micromonospora sp. SCSIO 07395. Molecules 2023, 28, 821. [Google Scholar] [CrossRef]
- Zhou, Q.; Luo, G.C.; Zhang, H.; Tang, G.L. Discovery of 16-Demethylrifamycins by Removing the Predominant Polyketide Biosynthesis Pathway in Micromonospora sp. Strain TP-A0468. Appl. Environ. Microbiol. 2019, 85, e02597-18. [Google Scholar] [CrossRef] [Green Version]
- Lasch, C.; Gummerlich, N.; Myronovskyi, M.; Palusczak, A.; Zapp, J.; Luzhetskyy, A. Loseolamycins: A Group of New Bioactive Alkylresorcinols Produced after Heterologous Expression of a Type III PKS from Micromonospora endolithica. Molecules 2020, 25, 4594. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wen, Z.; Liu, L.; Zhu, X.; Shen, B.; Yan, X.; Duan, Y.; Huang, Y. Yangpumicins F and G, Enediyne Congeners from Micromonospora yangpuensis DSM 45577. J. Nat. Prod. 2019, 82, 2483–2488. [Google Scholar] [CrossRef] [PubMed]
- Shi, T.; Li, Y.-J.; Wang, Z.-M.; Wang, Y.-F.; Wang, B.; Shi, D.-Y. New Pyrroline Isolated from Antarctic Krill-Derived Actinomycetes Nocardiopsis sp. LX-1 Combining with Molecular Networking. Mar. Drugs 2023, 21, 127. [Google Scholar] [CrossRef] [PubMed]
- Siddharth, S.; Aswathanarayan, J.B.; Kuruburu, M.G.; Madhunapantula, S.R.V.; Vittal, R.R. Diketopiperazine derivative from marine actinomycetes Nocardiopsis sp. SCA30 with antimicrobial activity against MRSA. Arch. Microbiol. 2021, 203, 6173–6181. [Google Scholar] [CrossRef]
- Braña, A.F.; Sarmiento-Vizcaíno, A.; Pérez-Victoria, I.; Otero, L.; Fernández, J.; Palacios, J.J.; Martín, J.; De La Cruz, M.; Díaz, C.; Vicente, F.; et al. Branimycins B and C, Antibiotics Produced by the Abyssal Actinobacterium Pseudonocardia carboxydivorans M-227. J. Nat. Prod. 2017, 80, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Matroodi, S.; Siitonen, V.; Baral, B.; Yamada, K.; Akhgari, A.; Metsä-Ketelä, M. Genotyping-Guided Discovery of Persiamycin A From Sponge-Associated Halophilic Streptomonospora sp. PA3. Front Microbiol. 2020, 11, 1237. [Google Scholar] [CrossRef]
- Pereira, F.; Almeida, J.R.; Paulino, M.; Grilo, I.R.; Macedo, H.; Cunha, I.; Sobral, R.G.; Vasconcelos, V.; Gaudêncio, S.P. Antifouling Napyradiomycins from Marine-Derived Actinomycetes Streptomyces aculeolatus. Mar. Drugs 2020, 18, 63. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Zhang, C.; Liu, Y.; Huang, Y.; Bai, Y.; Hang, X.; Zeng, L.; Zhu, D.; Bi, H. Armeniaspirol A: A novel anti-Helicobacter pylori agent. Microb. Biotechnol. 2022, 15, 442–454. [Google Scholar] [CrossRef]
- Rodríguez Estévez, M.; Gummerlich, N.; Myronovskyi, M.; Zapp, J.; Luzhetskyy, A. Benzanthric Acid, a Novel Metabolite From Streptomyces albus Del14 Expressing the Nybomycin Gene Cluster. Front. Chem. 2020, 7, 896. [Google Scholar] [CrossRef] [PubMed]
- Manikkam, R.; Murthy, S.; Palaniappan, S.; Kaari, M.; Sahu, A.K.; Said, M.; Ganesan, V.; Kannan, S.; Ramasamy, B.; Thirugnanasambandan, S. Antibacterial and Anti-HIV Metabolites from Marine Streptomyces albus MAB56 Isolated from Andaman and Nicobar Islands, India. Appl. Biochem. Biotechnol. 2023, 195, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.; Kwon, O.S.; Shin, J.; Oh, K.B. Antibacterial Activity and Mode of Action of Lactoquinomycin A from Streptomyces bacillaris. Mar. Drugs 2020, 19, 7. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Isolation and Characterization of a New Endophytic Actinobacterium Streptomyces californicus Strain ADR1 as a Promising Source of Anti-Bacterial, Anti-Biofilm and Antioxidant Metabolites. Microorganisms 2020, 8, 929. [Google Scholar] [CrossRef] [PubMed]
- Shaala, L.A.; Youssef, D.T.A.; Alzughaibi, T.A.; Elhady, S.S. Antimicrobial Chlorinated 3-Phenylpropanoic Acid Derivatives from the Red Sea Marine Actinomycete Streptomyces coelicolor LY001. Mar. Drugs 2020, 18, 450. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Chen, N.; Li, J.; Su, J.C.; Yang, J.; Zhang, C.X.; Lin, H.W.; Zhou, Y. Antimicrobial Chlorinated Carbazole Alkaloids from the Sponge-Associated Actinomycete Streptomyces diacarni LHW51701. Chin. J. Chem. 2021, 39, 1188–1192. [Google Scholar] [CrossRef]
- Takehana, Y.; Umekita, M.; Hatano, M.; Kato, C.; Sawa, R.; Igarashi, M. Fradiamine A, a new siderophore from the deep-sea actinomycete Streptomyces fradiae MM456M-mF7. J. Antibiot. 2017, 70, 611–615. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Ou, P.; Liu, L.; Jin, X. Anti-MRSA Activity of Actinomycin X2 and Collismycin A Produced by Streptomyces globisporus WA5-2-37 From the Intestinal Tract of American Cockroach (Periplaneta americana). Front. Microbiol. 2020, 11, 555. [Google Scholar] [CrossRef] [Green Version]
- Kaweewan, I.; Komaki, H.; Hemmi, H.; Kodani, S. Isolation and structure determination of a new thiopeptide globimycin from Streptomyces globisporus subsp. globisporus based on genome mining. Tetrahedron Lett. 2018, 59, 409–414. [Google Scholar] [CrossRef]
- Saleem, M.; Hassan, A.; Li, F.; Lu, Q.; Ponomareva, L.V.; Parkin, S.; Sun, C.; Thorson, J.S.; Shaaban, K.A.; Sajid, I. Bioprospecting of desert actinobacteria with special emphases on griseoviridin, mitomycin C and a new bacterial metabolite producing Streptomyces sp. PU-KB10–4. BMC Microbiol. 2023, 23, 69. [Google Scholar] [CrossRef]
- Harunari, E.; Imada, C.; Igarashi, Y. Konamycins A and B and Rubromycins CA1 and CA2, Aromatic Polyketides from the Tunicate-Derived Streptomyces hyaluromycini MB-PO13T. J. Nat. Prod. 2019, 82, 1609–1615. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Song, Y.; Li, X.; Wang, X.; Ling, C.; Qin, X.; Zhou, Z.; Li, Q.; Wei, X.; Ju, J. Abyssomicin Monomers and Dimers from the Marine-Derived Streptomyces koyangensis SCSIO 5802. J. Nat. Prod. 2018, 81, 1892–1898. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.; Kwon, O.S.; Chung, B.; Lee, J.; Sun, J.; Shin, J.; Oh, K.B. Antibacterial Activity of Chromomycins from a Marine-Derived Streptomyces microflavus. Mar. Drugs 2020, 18, 522. [Google Scholar] [CrossRef]
- Martinet, L.; Naômé, A.; Rezende, L.C.; Tellatin, D.; Pignon, B.; Docquier, J.-D.; Sannio, F.; Baiwir, D.; Mazzucchelli, G.; Frédérich, M. Lunaemycins, new cyclic hexapeptide antibiotics from the cave moonmilk-dweller Streptomyces lunaelactis MM109T. Int. J. Mol. Sci. 2023, 24, 1114. [Google Scholar] [CrossRef]
- Yang, L.; Hou, L.; Li, H.; Li, W. Antibiotic angucycline derivatives from the deepsea-derived Streptomyces lusitanus. Nat. Prod. Res. 2020, 34, 3444–3450. [Google Scholar] [CrossRef] [PubMed]
- Sujarit, K.; Mori, M.; Dobashi, K.; Shiomi, K.; Pathom-Aree, W.; Lumyong, S. New Antimicrobial Phenyl Alkenoic Acids Isolated from an Oil Palm Rhizosphere-Associated Actinomycete, Streptomyces palmae CMU-AB204T. Microorganisms 2020, 8, 350. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.; Tian, E.; Kong, F.; Hong, K. Bioactive Molecules from Mangrove Streptomyces qinglanensis 172205. Mar. Drugs 2020, 18, 255. [Google Scholar] [CrossRef]
- Heo, C.-S.; Kang, J.S.; Kwon, J.-H.; Anh, C.V.; Shin, H.J. Pyrrole-Containing Alkaloids from a Marine-Derived Actinobacterium Streptomyces zhaozhouensis and Their Antimicrobial and Cytotoxic Activities. Mar. Drugs 2023, 21, 167. [Google Scholar] [CrossRef] [PubMed]
- Bo, S.T.; Xu, Z.F.; Yang, L.; Cheng, P.; Tan, R.X.; Jiao, R.H.; Ge, H.M. Structure and biosynthesis of mayamycin B, a new polyketide with antibacterial activity from Streptomyces sp. 120454. J. Antibiot. 2018, 71, 601–605. [Google Scholar] [CrossRef]
- Liang, Y.; Xie, X.; Chen, L.; Yan, S.; Ye, X.; Anjum, K.; Huang, H.; Lian, X.; Zhang, Z. Bioactive Polycyclic Quinones from Marine Streptomyces sp. 182SMLY. Mar. Drugs 2016, 14, 10. [Google Scholar] [CrossRef] [Green Version]
- Safaei, N.; Mast, Y.; Steinert, M.; Huber, K.; Bunk, B.; Wink, J. Angucycline-like Aromatic Polyketide from a Novel Streptomyces Species Reveals Freshwater Snail Physa acuta as Underexplored Reservoir for Antibiotic-Producing Actinomycetes. Antibiotics 2020, 10, 22. [Google Scholar] [CrossRef]
- Wang, T.; Li, F.; Lu, Q.; Wu, G.; Jiang, Z.; Liu, S.; Habden, X.; Razumova, E.A.; Osterman, I.A.; Sergiev, P.V.; et al. Diversity, novelty, antimicrobial activity, and new antibiotics of cultivable endophytic actinobacteria isolated from psammophytes collected from Taklamakan Desert. J. Pharm. Anal. 2021, 11, 241–250. [Google Scholar] [CrossRef]
- Guerrero-Garzón, J.F.; Zehl, M.; Schneider, O.; Rückert, C.; Busche, T.; Kalinowski, J.; Bredholt, H.; Zotchev, S.B. Streptomyces spp. From the Marine Sponge Antho dichotoma: Analyses of Secondary Metabolite Biosynthesis Gene Clusters and Some of Their Products. Front. Microbiol. 2020, 11, 437. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Wang, X.; Huang, T.; Deng, Z.; Lin, S. Naphthoquinone-Based Meroterpenoids from Marine-Derived Streptomyces sp. B9173. Biomolecules 2020, 10, 1187. [Google Scholar] [CrossRef]
- Carretero-Molina, D.; Ortiz-López, F.J.; Martín, J.; Oves-Costales, D.; Díaz, C.; De La Cruz, M.; Cautain, B.; Vicente, F.; Genilloud, O.; Reyes, F. New Napyradiomycin Analogues from Streptomyces sp. Strain CA-271078. Mar. Drugs 2019, 18, 22. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Hu, X.; Hu, X.; Li, S.; Li, L.; Yu, L.; Liu, H.; You, X.; Wang, Z.; Li, L.; et al. Cytotoxic and Antibacterial Cervinomycins B1-4 from a Streptomyces Species. J. Nat. Prod. 2019, 82, 2337–2342. [Google Scholar] [CrossRef] [PubMed]
- Dardić, D.; Lauro, G.; Bifulco, G.; Laboudie, P.; Sakhaii, P.; Bauer, A.; Vilcinskas, A.; Hammann, P.E.; Plaza, A. Svetamycins A-G, Unusual Piperazic Acid-Containing Peptides from Streptomyces sp. J. Org. Chem. 2017, 82, 6032–6043. [Google Scholar] [CrossRef] [PubMed]
- Lü, Y.; Shao, M.; Wang, Y.; Qian, S.; Wang, M.; Wang, Y.; Li, X.; Bao, Y.; Deng, C.; Yue, C.; et al. Zunyimycins B and C, New Chloroanthrabenzoxocinones Antibiotics against Methicillin-Resistant Staphylococcus aureus and Enterococci from Streptomyces sp. FJS31-2. Molecules 2017, 22, 251. [Google Scholar] [CrossRef] [Green Version]
- Kitani, S.; Ueguchi, T.; Igarashi, Y.; Leetanasaksakul, K.; Thamchaipenet, A.; Nihira, T. Rakicidin F, a new antibacterial cyclic depsipeptide from a marine sponge-derived Streptomyces sp. J. Ant. 2017, 71, 139–141. [Google Scholar] [CrossRef]
- Yang, Z.; Shao, L.; Wang, M.; Rao, M.; Ge, M.; Xu, Y. Two novel quinomycins discovered by UPLC-MS from Stretomyces sp. HCCB11876. J. Ant. 2019, 72, 164–168. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.X.; Ding, R.; Jiang, S.T.; Tang, J.S.; Hu, D.; Chen, G.D.; Lin, F.; Hong, K.; Yao, X.S.; Gao, H. Aldgamycins J-O, 16-Membered Macrolides with a Branched Octose Unit from Streptomycetes sp. and Their Antibacterial Activities. J. Nat. Prod. 2016, 79, 2446–2454. [Google Scholar] [CrossRef]
- Zhou, B.; Ji, Y.Y.; Zhang, H.J.; Shen, L. Gephyyamycin and cysrabelomycin, two new angucyclinone derivatives from the Streptomyces sp. HN-A124. Nat. Prod. Res. 2021, 35, 2117–2122. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Qi, H.; Zhang, H.; Zhang, S.-Y.; Zhang, C.-H.; Zhang, L.-Q.; Xiang, W.-S.; Wang, J.-D. Anulamycins A–F, Cinnamoyl-Containing Peptides from a Lake Sediment Derived Streptomyces. J. Nat. Prod. 2023, 86, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, L.; Zhang, X.; Liang, Y.; Anjum, K.; Chen, L.; Lian, X.Y. Bioactive Bafilomycins and a New N-Arylpyrazinone Derivative from Marine-derived Streptomyces sp. HZP-2216E. Planta Med. 2017, 83, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Voitsekhovskaia, I.; Paulus, C.; Dahlem, C.; Rebets, Y.; Nadmid, S.; Zapp, J.; Axenov-Gribanov, D.; Rückert, C.; Timofeyev, M.; Kalinowski, J.; et al. Baikalomycins A-C, New Aquayamycin-Type Angucyclines Isolated from Lake Baikal Derived Streptomyces sp. IB201691-2A. Microorganisms 2020, 8, 680. [Google Scholar] [CrossRef]
- Iniyan, A.M.; Sudarman, E.; Wink, J.; Kannan, R.R.; Vincent, S.G.P. Ala-geninthiocin, a new broad spectrum thiopeptide antibiotic, produced by a marine Streptomyces sp. ICN19. J. Antibiot. 2019, 72, 99–105. [Google Scholar] [CrossRef]
- Son, S.; Hong, Y.S.; Jang, M.; Heo, K.T.; Lee, B.; Jang, J.P.; Kim, J.W.; Ryoo, I.J.; Kim, W.G.; Ko, S.K.; et al. Genomics-Driven Discovery of Chlorinated Cyclic Hexapeptides Ulleungmycins A and B from a Streptomyces Species. J. Nat. Prod. 2017, 80, 3025–3031. [Google Scholar] [CrossRef]
- Son, S.; Ko, S.K.; Kim, S.M.; Kim, E.; Kim, G.S.; Lee, B.; Ryoo, I.J.; Kim, W.G.; Lee, J.S.; Hong, Y.S.; et al. Antibacterial Cyclic Lipopeptide Enamidonins with an Enamide-Linked Acyl Chain from a Streptomyces Species. J. Nat. Prod. 2018, 81, 2462–2469. [Google Scholar] [CrossRef]
- Sawa, R.; Kubota, Y.; Umekita, M.; Hatano, M.; Hayashi, C.; Igarashi, M. Quadoctomycin, a 48-membered macrolide antibiotic from Streptomyces sp. MM168-141F8. J. Antibiot. 2017, 71, 91–96. [Google Scholar] [CrossRef]
- Konwar, A.N.; Basak, S.; Devi, S.G.; Borah, J.C.; Thakur, D. Streptomyces sp. MNP32, a forest-dwelling Actinomycetia endowed with potent antibacterial metabolites. 3 Biotech 2023, 13, 257. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, J.; Liu, C.L.; Xiang, L.; Ma, S.Y.; Li, W.; Jiao, R.H.; Tan, R.X.; Ge, H.M. New borrelidin derivatives from an endophytic Streptomyces sp. Tetrahedron Lett. 2018, 59, 4517–4520. [Google Scholar] [CrossRef]
- Guo, Z.K.; Wang, Y.C.; Tan, Y.Z.; Abulaizi, A.; Xiong, Z.J.; Zhang, S.Q.; Yang, Y.; Yang, L.Y.; Shi, J. Nagimycins A and B, Antibacterial Ansamycin-Related Macrolactams from Streptomyces sp. NA07423. Org. Lett. 2023, 25, 4203–4207. [Google Scholar] [CrossRef] [PubMed]
- Arn, F.; Frasson, D.; Kroslakova, I.; Rezzonico, F.; Pothie, J.F.; Riedl, R.; Sievers, M. Isolation and Identification of Actinomycetes Strains from Switzerland and their Biotechnological Potential. Chimia 2020, 74, 382–390. [Google Scholar] [CrossRef]
- Mazumdar, R.; Dutta, P.P.; Saikia, J.; Borah, J.C.; Thakur, D. Streptomyces sp. Strain PBR11, a Forest-Derived Soil Actinomycetia with Antimicrobial Potential. Microbiol. Spectr. 2023, 11, 17544. [Google Scholar] [CrossRef]
- Betancur, L.A.; Forero, A.M.; Vinchira-Villarraga, D.M.; Cárdenas, J.D.; Romero-Otero, A.; Chagas, F.O.; Pupo, M.T.; Castellanos, L.; Ramos, F.A. NMR-based metabolic profiling to follow the production of anti-phytopathogenic compounds in the culture of the marine strain Streptomyces sp. PNM-9. Microbiol. Res. 2020, 239, 126507. [Google Scholar] [CrossRef]
- Cheng, C.; Othman, E.M.; Reimer, A.; Grüne, M.; Kozjak-Pavlovic, V.; Stopper, H.; Hentschel, U.; Abdelmohsen, U.R. Ageloline A, new antioxidant and antichlamydial quinolone from the marine sponge-derived bacterium Streptomyces sp. SBT345. Tetrahedron Lett. 2016, 57, 2786–2789. [Google Scholar] [CrossRef]
- Cong, Z.; Huang, X.; Liu, Y.; Liu, Y.; Wang, P.; Liao, S.; Yang, B.; Zhou, X.; Huang, D.; Wang, J. Cytotoxic anthracycline and antibacterial tirandamycin analogues from a marine-derived Streptomyces sp. SCSIO 41399. J. Antibiot. 2019, 72, 45–49. [Google Scholar] [CrossRef]
- Tian, H.; Shafi, J.; Ji, M.; Bi, Y.; Yu, Z. Antimicrobial Metabolites from Streptomyces sp. SN0280. J. Nat. Prod. 2017, 80, 1015–1019. [Google Scholar] [CrossRef]
- Maiti, P.K.; Das, S.; Sahoo, P.; Mandal, S. Streptomyces sp. SM01 isolated from Indian soil produces a novel antibiotic picolinamycin effective against multi drug resistant bacterial strains. Sci. Rep. 2020, 10, 788. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, X.; He, N.; Xie, Y.; Hong, B. Rescrutiny of the sansanmycin biosynthetic gene cluster leads to the discovery of a novel sansanmycin analogue with more potency against Mycobacterium tuberculosis. J. Antibiot. 2019, 72, 769–774. [Google Scholar] [CrossRef]
- Pratiwi, R.H.; Hidayat, I.; Hanafi, M.; Mangunwardoyo, W. Isolation and structure elucidation of phenazine derivative from Streptomyces sp. strain UICC B-92 isolated from Neesia altissima (Malvaceae). Iran. J. Microbiol. 2020, 12, 127. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Zhang, D.S.; Zhang, H.J.; Li, J.Q.; Ding, W.J.; Xu, C.D.; Ma, Z.J. Medermycin-Type Naphthoquinones from the Marine-Derived Streptomyces sp. XMA39. J. Nat. Prod. 2018, 81, 2120–2124. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ren, Z.; Chunyu, W.X.; Li, G.D.; Chen, X.; Zhang, Z.T.L.; Sun, H.B.; Wang, M.; Xie, T.P.; Wang, M.; et al. Exploration of Diverse Secondary Metabolites From Streptomyces sp. YINM00001, Using Genome Mining and One Strain Many Compounds Approach. Front. Microbiol. 2022, 13, 831174. [Google Scholar] [CrossRef] [PubMed]
- Newaz, A.W.; Yong, K.; Lian, X.Y.; Zhang, Z. Streptoindoles A–D, novel antimicrobial indole alkaloids from the marine-associated actinomycete Streptomyces sp. ZZ1118. Tetrahedron 2022, 104, 132598. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, W.; Ge, H.; Zhang, Z.; Wu, B. Bioactive Streptoglutarimides A-J from the Marine-Derived Streptomyces sp. ZZ741. J. Nat. Prod. 2019, 82, 2800–2808. [Google Scholar] [CrossRef] [PubMed]
- Asolkar, R.N.; Singh, A.; Jensen, P.R.; Aalbersberg, W.; Carté, B.K.; Feussner, K.D.; Subramani, R.; DiPasquale, A.; Rheingold, A.L.; Fenical, W. Marinocyanins, cytotoxic bromo-phenazinone meroterpenoids from a marine bacterium from the streptomycete clade MAR4. Tetrahedron 2017, 73, 2234–2241. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Kagoshima, Y.; Masuda, T.; Kizuka, M.; Ogawa, Y.; Endo, S.; Nishigoori, H.; Saito, K.; Takasugi, K.; Miura, M.; et al. Muraminomicins, new lipo-nucleoside antibiotics from Streptosporangium sp. SANK 60501-structure elucidations of muraminomicins and supply of the core component for derivatization. J. Antibiot. 2019, 72, 943–955. [Google Scholar] [CrossRef]
- Teta, R.; Marteinsson, V.T.; Longeon, A.; Klonowski, A.M.; Groben, R.; Bourguet-Kondracki, M.L.; Costantino, V.; Mangoni, A. Thermoactinoamide A, an Antibiotic Lipophilic Cyclopeptide from the Icelandic Thermophilic Bacterium Thermoactinomyces vulgaris. J. Nat. Prod. 2017, 80, 2530–2535. [Google Scholar] [CrossRef]
- Zhang, S.; Xie, Q.; Sun, C.; Tian, X.P.; Gui, C.; Qin, X.; Zhang, H.; Ju, J. Cytotoxic Kendomycins Containing the Carbacylic Ansa Scaffold from the Marine-Derived Verrucosispora sp. SCSIO 07399. J. Nat. Prod. 2019, 82, 3366–3371. [Google Scholar] [CrossRef]
- Gupte, M.; Kulkarni, P.; Ganguli, B.N. Antifungal antibiotics. Appl. Microbiol. Biotechnol. 2002, 58, 46–57. [Google Scholar] [CrossRef]
- Hazen, E.L.; Brown, R. Fungicidin, an Antibiotic Produced by a Soil Actinomycete. Proc. Soc. Exp. Biol. Med. 1951, 76, 93–97. [Google Scholar] [CrossRef]
- Georgopapadakou, N.H. Antifungals: Mechanism of action and resistance, established and novel drugs. Curr. Opin. Microbiol. 1998, 1, 547–557. [Google Scholar] [CrossRef]
- Ghannoum, M.A.; Rice, L.B. Antifungal agents: Mode of action, mechanisms of resistance, and correlation of these mechanisms with bacterial resistance. Clin. Microbiol. Rev. 1999, 12, 501–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Intra, B.; Euanorasetr, J.; Nihira, T.; Panbangred, W. Characterization of a gamma-butyrolactone synthetase gene homologue (stcA) involved in bafilomycin production and aerial mycelium formation in Streptomyces sp. SBI034. Appl. Microbiol. Biotechnol. 2016, 100, 2749–2760. [Google Scholar] [CrossRef] [PubMed]
- Intra, B.; Greule, A.; Bechthold, A.; Euanorasetr, J.; Paululat, T.; Panbangred, W. Thailandins A and B, New Polyene Macrolactone Compounds Isolated from Actinokineospora bangkokensis Strain 44EHWT, Possessing Antifungal Activity against Anthracnose Fungi and Pathogenic Yeasts. J. Agric. Food Chem. 2016, 64, 5171–5179. [Google Scholar] [CrossRef] [PubMed]
- Euanorasetr, J.; Junhom, M.; Tantimavanich, S.; Vorasin, O.; Munyoo, B.; Tuchinda, P.; Panbangred, W. Halogenated benzoate derivatives of altholactone with improved anti-fungal activity. J. Asian Nat. Prod. Res. 2016, 18, 462–474. [Google Scholar] [CrossRef]
- Bunyapaiboonsri, T.; Yoiprommarat, S.; Suriyachadkun, C.; Supothina, S.; Chanthaket, R.; Chutrakul, C.; Vichai, V. Actinomadurone, a polycyclic tetrahydroxanthone from Actinomadura sp. BCC 35430. Tetrahedron Lett. 2017, 58, 3223–3225. [Google Scholar] [CrossRef]
- Cheng, M.-J.; Chen, J.-J.; Wu, M.-D.; Leu, J.-Y.; Tseng, M. Antifungal Activities of Compounds Produced by Newly Isolated Acrocarpospora Strains. Antibiotics 2023, 12, 95. [Google Scholar] [CrossRef]
- Kim, H.R.; Kim, J.; Yu, J.S.; Lee, B.S.; Kim, K.H.; Kim, C.S. Isolation, structure elucidation, total synthesis, and biosynthesis of dermazolium A, an antibacterial imidazolium metabolite of a vaginal bacterium Dermabacter vaginalis. Arch. Pharm. Res. 2023, 46, 35–43. [Google Scholar] [CrossRef]
- Hara, S.; Ishikawa, N.; Hara, Y.; Nehira, T.; Sakai, K.; Gonoi, T.; Ishibashi, M. Nabscessins A and B, Aminocyclitol Derivatives from Nocardia abscessus IFM 10029T. J. Nat. Prod. 2017, 80, 565–568. [Google Scholar] [CrossRef]
- Kim, H.J.; Han, C.Y.; Park, J.S.; Oh, S.H.; Kang, S.H.; Choi, S.S.; Kim, J.M.; Kwak, J.H.; Kim, E.S. Nystatin-like Pseudonocardia polyene B1, a novel disaccharide-containing antifungal heptaene antibiotic. Sci. Rep. 2018, 8, 13584. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Gao, X.; Kang, Z.; Huang, L.; Fan, D.; Yan, X. Saccharothrix yanglingensis Strain Hhs.015 Is a Promising Biocontrol Agent on Apple Valsa Canker. Plant Dis. 2016, 100, 510–514. [Google Scholar] [CrossRef] [Green Version]
- Thekkangil, A.; George, B.; Prakash, S.M.U.; Suchithra, T.V. Mechanism of Streptomyces albidoflavus STV1572a derived 1-heneicosanol as an inhibitor against squalene epoxidase of Trichophyton mentagrophytes. Microb. Pathog. 2021, 154, 104853. [Google Scholar] [CrossRef]
- Ding, N.; Jiang, Y.; Han, L.; Chen, X.; Ma, J.; Qu, X.; Mu, Y.; Liu, J.; Li, L.; Jiang, C.; et al. Bafilomycins and Odoriferous Sesquiterpenoids from Streptomyces albolongus Isolated from Elephas maximus Feces. J. Nat. Prod. 2016, 79, 799–805. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Fu, S.N.; Bao, Y.X.; Yang, Y.; Shen, H.F.; Lin, B.R.; Zhou, G.X. Kitamycin C, a new antimycin-type antibiotic from Streptomyces antibioticus strain 200-09. Nat. Prod. Res. 2017, 31, 1819–1824. [Google Scholar] [CrossRef] [PubMed]
- Pérez, M.; Schleissner, C.; Fernández, R.; Rodríguez, P.; Reyes, F.; Zuñiga, P.; De La Calle, F.; Cuevas, C. PM100117 and PM100118, new antitumor macrolides produced by a marine Streptomyces caniferus GUA-06-05-006A. J. Antibiot. 2016, 69, 388–394. [Google Scholar] [CrossRef] [PubMed]
- Vicente Dos Reis, G.; Abraham, W.R.; Grigoletto, D.F.; De Campos, J.B.; Marcon, J.; Da Silva, J.A.; Quecine, M.C.; De Azevedo, J.L.; Ferreira, A.G.; De Lira, S.P. Gloeosporiocide, a new antifungal cyclic peptide from Streptomyces morookaense AM25 isolated from the Amazon bulk soil. FEMS Microbiol. Lett. 2019, 366, 175. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yu, Z.; Zhao, J.; Zhuang, X.; Cao, P.; Guo, X.; Liu, C.; Xiang, W. Community Composition, Antifungal Activity and Chemical Analyses of Ant-Derived Actinobacteria. Front. Microbiol. 2020, 11, 201. [Google Scholar] [CrossRef] [Green Version]
- Mojicevic, M.; D’Agostino, P.M.; Pavic, A.; Vojnovic, S.; Senthamaraikannan, R.; Vasiljevic, B.; Gulder, T.A.M.; Nikodinovic-Runic, J. Streptomyces sp. BV410 isolate from chamomile rhizosphere soil efficiently produces staurosporine with antifungal and antiangiogenic properties. Microbiol. Open 2020, 9, e986. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Peng, D.; Peng, F.-F.; Zhang, Q.-B.; Duan, Y.-W.; Huang, Y. The Isolation and Structure Elucidation of Spirotetronate Lobophorins A, B, and H8 from Streptomyces sp. CB09030 and Their Biosynthetic Gene Cluster. Molecules 2023, 28, 3597. [Google Scholar] [CrossRef]
- Fang, Q.; Maglangit, F.; Mugat, M.; Urwald, C.; Kyeremeh, K.; Deng, H. Targeted Isolation of Indole Alkaloids from Streptomyces sp. CT37. Molecules 2020, 25, 1108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.; Niu, H.J.; Qu, T.L.; Zhang, X.F.; Du, F.Y. Streptomyces sp. FX13 inhibits fungicide-resistant Botrytis cinerea in vitro and in vivo by producing oligomycin A. Pestic. Biochem. Physiol. 2021, 175, 104834. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Li, C.; Wang, H.; Yu, Z.; Xu, X.; Wang, X.; Zhao, J.; Xiang, W. Community Structures and Antifungal Activity of Root-Associated Endophytic Actinobacteria in Healthy and Diseased Cucumber Plants and Streptomyces sp. HAAG3-15 as a Promising Biocontrol Agent. Microorganisms 2020, 8, 236. [Google Scholar] [CrossRef] [Green Version]
- Yu, Z.; Wang, L.; Yang, J.; Zhang, F.; Sun, Y.; Yu, M.; Yan, Y.; Ma, Y.T.; Huang, S.X. A new antifungal macrolide from Streptomyces sp. KIB-H869 and structure revision of halichomycin. Tetrahedron Lett. 2016, 57, 1375–1378. [Google Scholar] [CrossRef]
- Feng, X.-L.; Zhang, R.-Q.; Wang, D.-C.; Dong, W.-G.; Wang, Z.-X.; Zhai, Y.-J.; Han, W.-B.; Yin, X.; Tian, J.; Wei, J. Genomic and Metabolite Profiling Reveal a Novel Streptomyces Strain, QHH-9511, from the Qinghai-Tibet Plateau. Microbiol. Spectr. 2023, 11, e02764-02722. [Google Scholar] [CrossRef] [PubMed]
- Yi, W.; Qin, L.; Lian, X.Y.; Zhang, Z. New Antifungal Metabolites from the Mariana Trench Sediment-Associated Actinomycete Streptomyces sp. SY1965. Mar. Drugs 2020, 18, 385. [Google Scholar] [CrossRef]
- Herbrík, A.; Corretto, E.; Chroňáková, A.; Langhansová, H.; Petrásková, P.; Hrdý, J.; Čihák, M.; Krištůfek, V.; Bobek, J.; Petříček, M.; et al. A Human Lung-Associated Streptomyces sp. TR1341 Produces Various Secondary Metabolites Responsible for Virulence, Cytotoxicity and Modulation of Immune Response. Front. Microbiol. 2020, 10, 3028. [Google Scholar] [CrossRef] [Green Version]
- Dutta, J.; Thakur, D. Evaluation of Antagonistic and Plant Growth Promoting Potential of Streptomyces sp. TT3 Isolated from Tea (Camellia sinensis) Rhizosphere Soil. Curr. Microbiol. 2020, 77, 1829–1838. [Google Scholar] [CrossRef]
- Yamamoto, K.; Futamura, Y.; Uson-Lopez, R.A.; Aono, H.; Shimizu, T.; Osada, H. YO-001A, a new antifungal agent produced by Streptomyces sp. YO15-A001. J. Antibiot. 2019, 72, 986–990. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Sharma, R.; Srinivas, V.; Naresh, N.; Mishra, S.P.; Ankati, S.; Pratyusha, S.; Govindaraj, M.; Gonzalez, S.V.; Nervik, S.; et al. Identification and Characterization of a Streptomyces albus Strain and Its Secondary Metabolite Organophosphate against Charcoal Rot of Sorghum. Plants 2020, 9, 1727. [Google Scholar] [CrossRef]
- Hoshino, S.; Wong, C.P.; Ozeki, M.; Zhang, H.; Hayashi, F.; Awakawa, T.; Asamizu, S.; Onaka, H.; Abe, I. Umezawamides, new bioactive polycyclic tetramate macrolactams isolated from a combined-culture of Umezawaea sp. and mycolic acid-containing bacterium. J. Antibiot. 2018, 71, 653–657. [Google Scholar] [CrossRef]
- Barreiro, C.; Prieto, C.; Sola-Landa, A.; Solera, E.; Martínez-Castro, M.; Pérez-Redondo, R.; García-Estrada, C.; Aparicio, J.F.; Fernández-Martínez, L.T.; Santos-Aberturas, J.; et al. Draft genome of Streptomyces tsukubaensis NRRL 18488, the producer of the clinically important immunosuppressant tacrolimus (FK506). J. Bacteriol. 2012, 194, 3756–3757. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bierer, B.E.; Mattila, P.S.; Standaert, R.F.; Herzenberg, L.A.; Burakoff, S.J.; Crabtree, G.; Schreiber, S.L. Two distinct signal transmission pathways in T lymphocytes are inhibited by complexes formed between an immunophilin and either FK506 or rapamycin. Proc. Nat. Acad. Sci. USA 1990, 87, 9231–9235. [Google Scholar] [CrossRef] [PubMed]
- Ohta, Y.; Ikeda, M. Deodorization of pig feces by actinomycetes. Appl. Environ. Microbiol. 1978, 36, 487–491. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Murata, A.; Shinsaku, H. Accelerated composting of cereal shochu-distillery wastes by actinomycetes. J. Ferment. Bioeng. 1995, 80, 421. [Google Scholar] [CrossRef]
- Mansour, F.A.; Mohamedin, A.H. Enzymes of Candida albicans cell-wall lytic system produced by Streptomyces thermodiastaticus. Acta Microbiol. Immunol. Hung. 2001, 48, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant Growth Promoting and Biocontrol Activity of Streptomyces spp. as Endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef] [Green Version]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.S.; Jeong, G.S. Salinosporamide A, a Marine-Derived Proteasome Inhibitor, Inhibits T Cell Activation through Regulating Proliferation and the Cell Cycle. Molecules 2020, 25, 5031. [Google Scholar] [CrossRef]
- Arcamone, F.; Cassinelli, G.; Fantini, G.; Grein, A.; Orezzi, P.; Pol, C.; Spalla, C. Adriamycin, 14-hydroxydaimomycin, a new antitumor antibiotic from S. peucetius var. caesius. Biotechnol. Bioeng. 1969, 11, 1101–1110. [Google Scholar] [CrossRef]
- Jagannathan, S.V.; Manemann, E.M.; Rowe, S.E.; Callender, M.C.; Soto, W. Marine Actinomycetes, New Sources of Biotechnological Products. Mar. Drugs 2021, 19, 365. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Shimasaki, R.; Miyanaga, S.; Oku, N.; Onaka, H.; Sakurai, H.; Saiki, I.; Kitani, S.; Nihira, T.; Wimonsiravude, W.; et al. Rakicidin D, an inhibitor of tumor cell invasion from marine-derived Streptomyces sp. J. Antibiot. 2010, 63, 563–565. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Chen, Y.; Ukaji, T.; Okada, S.; Umezawa, K. Isolation of ketomycin from Actinomycetes as an inhibitor of 2D and 3D cancer cell invasion. J. Antibiot. 2018, 72, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Choi, B.K.; Lee, H.S.; Kang, J.S.; Shin, H.J. Dokdolipids A−C, Hydroxylated Rhamnolipids from the Marine-Derived Actinomycete Actinoalloteichus hymeniacidonis. Mar. Drugs 2019, 17, 237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, T.; Iwatsuki, M.; Asami, Y.; Ishiyama, A.; Hokari, R.; Otoguro, K.; Matsumoto, A.; Sato, N.; Shiomi, K.; Takahashi, Y.; et al. Anti-trypanosomal compound, sagamilactam, a new polyene macrocyclic lactam from Actinomadura sp. K13-0306. J. Antibiot. 2016, 69, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Matsuoka, N.; In, Y.; Kataura, T.; Tashiro, E.; Saiki, I.; Sudoh, Y.; Duangmal, K.; Thamchaipenet, A. Nonthmicin, a Polyether Polyketide Bearing a Halogen-Modified Tetronate with Neuroprotective and Antiinvasive Activity from Actinomadura sp. Org. Lett. 2017, 19, 1406–1409. [Google Scholar] [CrossRef]
- Lu, C.; Xie, F.; Shan, C.; Shen, Y. Two novel cyclic hexapeptides from the genetically engineered Actinosynnema pretiosum. Appl. Microbiol. Biotechnol. 2017, 101, 2273–2279. [Google Scholar] [CrossRef]
- Frattaruolo, L.; Lacret, R.; Cappello, A.R.; Truman, A.W. A Genomics-Based Approach Identifies a Thioviridamide-Like Compound with Selective Anticancer Activity. ACS Chem. Biol. 2017, 12, 2815–2822. [Google Scholar] [CrossRef]
- Hoshino, S.; Ozeki, M.; Awakawa, T.; Morita, H.; Onaka, H.; Abe, I. Catenulobactins A and B, Heterocyclic Peptides from Culturing Catenuloplanes sp. with a Mycolic Acid-Containing Bacterium. J. Nat. Prod. 2018, 81, 2106–2110. [Google Scholar] [CrossRef]
- Pournejati, R.; Gust, R.; Kircher, B.; Karbalaei-Heidari, H.R. Microindoline 581, an Indole Derivative from Microbacterium Sp. RP581 as A Novel Selective Antineoplastic Agent to Combat Hepatic Cancer Cells: Production, Optimization and Structural Elucidation. Iran. J. Pharm. Res. 2020, 19, 290–305. [Google Scholar] [CrossRef]
- Fu, G.; Wang, R.; Ding, J.; Qi, H.; Zhao, Z.; Chen, C.; Zhang, H.; Xue, Z.; Wang, J.; Wu, M. Micromonospora zhangzhouensis sp. nov., a Novel Actinobacterium Isolated from Mangrove Soil, Exerts a Cytotoxic Activity in vitro. Sci. Rep. 2020, 10, 3889. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarmiento-Vizcaíno, A.; Braña, A.F.; Pérez-Victoria, I.; Martín, J.; De Pedro, N.; De La Cruz, M.; Díaz, C.; Vicente, F.; Acuña, J.L.; Reyes, F.; et al. Paulomycin G, a New Natural Product with Cytotoxic Activity against Tumor Cell Lines Produced by Deep-Sea Sediment Derived Micromonospora matsumotoense M-412 from the Avilés Canyon in the Cantabrian Sea. Mar. Drugs 2017, 15, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.J.; Zhang, S.Y.; Ye, Y.H.; Yu, Z.; Qi, H.; Zhang, H.; Xue, Z.L.; Wang, J.D.; Wu, M. Three New Isoflavonoid Glycosides from the Mangrove-Derived Actinomycete Micromonospora aurantiaca 110B. Mar. Drugs 2019, 17, 294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Zhao, W.; Jiang, H.L.; Zhou, J.; Chen, X.M.; Lian, Y.Y.; Jiang, H.; Lin, F. Rakicidins G-I, cyclic depsipeptides from marine Micromonospora chalcea FIM 02-523. Tetrahedron 2018, 74, 4151–4154. [Google Scholar] [CrossRef]
- Nie, Y.L.; Wu, Y.D.; Wang, C.X.; Lin, R.; Xie, Y.; Fang, D.S.; Jiang, H.; Lian, Y.Y. Structure elucidation and antitumour activity of a new macrolactam produced by marine-derived actinomycete Micromonospora sp. FIM05328. Nat. Prod. Res. 2018, 32, 2133–2138. [Google Scholar] [CrossRef]
- Gao, M.Y.; Qi, H.; Li, J.S.; Zhang, H.; Zhang, J.; Wang, J.D.; Xiang, W.S. A new naphthalenepropanoic acid analog from the marine-derived actinomycetes Micromonospora sp. HS-HM-036. J. Asian Nat. Prod. Res. 2017, 19, 930–934. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Chen, J.J.; Adhikari, A.; Yang, D.; Crnovcic, I.; Wang, N.; Chang, C.Y.; Rader, C.; Shen, B. Genome Mining of Micromonospora yangpuensis DSM 45577 as a Producer of an Anthraquinone-Fused Enediyne. Org. Lett. 2017, 19, 6192–6195. [Google Scholar] [CrossRef]
- Fukuda, T.; Takahashi, M.; Nagai, K.; Harunari, E.; Imada, C.; Tomoda, H. Isomethoxyneihumicin, a new cytotoxic agent produced by marine Nocardiopsis alba KM6-1. J. Antibiot. 2017, 70, 590–594. [Google Scholar] [CrossRef] [Green Version]
- Messaoudi, O.; Sudarman, E.; Bendahou, M.; Jansen, R.; Stadler, M.; Wink, J. Kenalactams A-E, Polyene Macrolactams Isolated from Nocardiopsis CG3. J. Nat. Prod. 2019, 82, 1081–1088. [Google Scholar] [CrossRef]
- Shaaban, K.A.; Shaaban, M.; Rahman, H.; Grün-Wollny, I.; Kämpfer, P.; Kelter, G.; Fiebig, H.H.; Laatsch, H. Karamomycins A-C: 2-Naphthalen-2-yl-thiazoles from Nonomuraea endophytica. J. Nat. Prod. 2019, 82, 870–877. [Google Scholar] [CrossRef]
- Yang, T.; Yamada, K.; Zhou, T.; Harunari, E.; Igarashi, Y.; Terahara, T.; Kobayashi, T.; Imada, C. Akazamicin, a cytotoxic aromatic polyketide from marine-derived Nonomuraea sp. J. Antibiot. 2019, 72, 202–209. [Google Scholar] [CrossRef] [PubMed]
- Gamaleldin, N.M.; Bakeer, W.; Sayed, A.M.; Shamikh, Y.I.; El-Gendy, A.O.; Hassan, H.M.; Horn, H.; Abdelmohsen, U.R.; Hozzein, W.N. Exploration of Chemical Diversity and Antitrypanosomal Activity of Some Red Sea-Derived Actinomycetes Using the OSMAC Approach Supported by LC-MS-Based Metabolomics and Molecular Modelling. Antibiotics 2020, 9, 629. [Google Scholar] [CrossRef] [PubMed]
- Anh, C.V.; Kwon, J.-H.; Kang, J.S.; Lee, H.-S.; Heo, C.-S.; Shin, H.J. New Angucycline Glycosides from a Marine-Derived Bacterium Streptomyces ardesiacus. Int. J. Mol. Sci. 2022, 23, 13779. [Google Scholar] [CrossRef] [PubMed]
- Kaari, M.; Joseph, J.; Manikkam, R.; Kalyanasundaram, R.; Sivaraj, A.; Anbalmani, S.; Murthy, S.; Sahu, A.K.; Said, M.; Dastager, S.G. A Novel Finding: 2, 4-Di-tert-butylphenol from Streptomyces bacillaris ANS2 Effective Against Mycobacterium tuberculosis and Cancer Cell Lines. Appl. Biochem. Biotechnol. 2023, 195, 1–14. [Google Scholar] [CrossRef]
- Gui, C.; Yuan, J.; Mo, X.; Huang, H.; Zhang, S.; Gu, Y.C.; Ju, J. Cytotoxic Anthracycline Metabolites from a Recombinant Streptomyces. J. Nat. Prod. 2018, 81, 1278–1289. [Google Scholar] [CrossRef]
- Kifer, D.; Mužinić, V.; Klaric, M.Š. Antimicrobial potency of single and combined mupirocin and monoterpenes, thymol, menthol and 1,8-cineole against Staphylococcus aureus planktonic and biofilm growth. J. Antibiot. 2016, 69, 689–696. [Google Scholar] [CrossRef]
- Xiao, F.; Li, H.; Xu, M.; Li, T.; Wang, J.; Sun, C.; Hong, K.; Li, W. Staurosporine Derivatives Generated by Pathway Engineering in a Heterologous Host and Their Cytotoxic Selectivity. J. Nat. Prod. 2018, 81, 1745–1751. [Google Scholar] [CrossRef]
- Kaweewan, I.; Komaki, H.; Hemmi, H.; Hoshino, K.; Hosaka, T.; Isokawa, G.; Oyoshi, T.; Kodani, S. Isolation and structure determination of a new cytotoxic peptide, curacozole, from Streptomyces curacoi based on genome mining. J. Antibiot. 2019, 72, 1–7. [Google Scholar] [CrossRef]
- Ortiz-López, F.J.; Alcalde, E.; Sarmiento-Vizcaíno, A.; Díaz, C.; Cautain, B.; García, L.A.; Blanco, G.; Reyes, F. New 3-Hydroxyquinaldic Acid Derivatives from Cultures of the Marine Derived Actinomycete Streptomyces cyaneofuscatus M-157. Mar. Drugs 2018, 16, 371. [Google Scholar] [CrossRef] [Green Version]
- Kimata, S.; Matsuda, T.; Suizu, Y.; Hayakawa, Y. Prodigiosin R2, a new prodigiosin from the roseophilin producer Streptomyces griseoviridis 2464-S5. J. Antibiot. 2018, 71, 393–396. [Google Scholar] [CrossRef]
- Liu, S.H.; Wang, W.; Wang, K.B.; Zhang, B.; Li, W.; Shi, J.; Jiao, R.H.; Tan, R.X.; Ge, H.M. Heterologous Expression of a Cryptic Giant Type I PKS Gene Cluster Leads to the Production of Ansaseomycin. Org. Lett. 2019, 21, 3785–3788. [Google Scholar] [CrossRef] [PubMed]
- Koomsiri, W.; Inahashi, Y.; Kimura, T.; Shiomi, K.; Takahashi, Y.; Omura, S.; Thamchaipenet, A.; Nakashima, T. Bisoxazolomycin A: A new natural product from ‘Streptomyces subflavus subsp. irumaensis’ AM-3603. J. Antibiot. 2017, 70, 1142–1145. [Google Scholar] [CrossRef]
- Zheng, D.; Ding, N.; Jiang, Y.; Zhang, J.; Ma, J.; Chen, X.; Liu, J.; Han, L.; Huang, X. Albaflavenoid, a new tricyclic sesquiterpenoid from Streptomyces violascens. J. Antibiot. 2016, 69, 773–775. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Han, L.; Qu, X.; Chen, X.; Zhong, J.; Bi, X.; Liu, J.; Jiang, Y.; Jiang, C.; Huang, X. Cytotoxic Fusicoccane-Type Diterpenoids from Streptomyces violascens Isolated from Ailuropoda melanoleuca Feces. J. Nat. Prod. 2017, 80, 837–844. [Google Scholar] [CrossRef]
- Lv, Q.; Fan, Y.; Tao, G.; Fu, P.; Zhai, J.; Ye, B.; Zhu, W. Sekgranaticin, a SEK34b-Granaticin Hybrid Polyketide from Streptomyces sp. 166. J. Org Chem. 2019, 84, 9087–9092. [Google Scholar] [CrossRef]
- Liang, Y.; Chen, L.; Ye, X.; Anjum, K.; Lian, X.Y.; Zhang, Z. New streptophenazines from marine Streptomyces sp. 182SMLY. Nat. Prod. Res. 2017, 31, 411–417. [Google Scholar] [CrossRef]
- Liu, C.X.; Liu, S.H.; Zhao, J.W.; Zhang, J.; Wang, X.J.; Li, J.S.; Wang, J.D.; Xiang, W.S. A new spectinabilin derivative with cytotoxic activity from ant-derived Streptomyces sp. 1H-GS5. J. Asian Nat. Prod. Res. 2017, 19, 924–929. [Google Scholar] [CrossRef]
- Zhou, B.; Qin, L.L.; Ding, W.J.; Ma, Z.J. Cytotoxic indolocarbazoles alkaloids from the Streptomyces sp. A65. Tetrahedron 2018, 74, 726–730. [Google Scholar] [CrossRef]
- Wang, X.; Elshahawi, S.I.; Ponomareva, L.V.; Ye, Q.; Liu, Y.; Copley, G.C.; Hower, J.C.; Hatcher, B.E.; Kharel, M.K.; Van Lanen, S.G.; et al. Structure Determination, Functional Characterization, and Biosynthetic Implications of Nybomycin Metabolites from a Mining Reclamation Site-Associated Streptomyces. J. Nat. Prod. 2019, 82, 3469–3476. [Google Scholar] [CrossRef]
- Saito, S.; Fujimaki, T.; Panbangred, W.; Sawa, R.; Igarashi, Y.; Imoto, M. Antarlides F–H, new members of the antarlide family produced by Streptomyces sp. BB47. J. Antibiot. 2017, 70, 595–600. [Google Scholar] [CrossRef] [Green Version]
- Han, X.; Liu, Z.; Zhang, Z.; Zhang, X.; Zhu, T.; Gu, Q.; Li, W.; Che, Q.; Li, D. Geranylpyrrol A and Piericidin F from Streptomyces sp. CHQ-64 ΔrdmF. J. Nat. Prod. 2017, 80, 1684–1687. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.N.; Zhang, H.J.; Li, J.Q.; Ding, W.J.; Ma, Z.J. Bioactive Indolocarbazoles from the Marine-Derived Streptomyces sp. DT-A61. J. Nat. Prod. 2018, 81, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, M.S.; Elmallah, M.I.Y.; Mohamed, A.A.; Ishibashi, M. Sharkquinone, a new ana-quinonoid tetracene derivative from marine-derived Streptomyces sp. EGY1 with TRAIL resistance-overcoming activity. J. Nat. Med. 2017, 71, 564–569. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Lu, M.C.; Chung, H.M.; Weng, C.F.; Su, J.H.; Yang, Y.T.; Su, Y.D.; Chang, Y.C.; Kuo, J.; Wu, Y.C.; et al. Bafilomycin M, a new cytotoxic bafilomycin produced by a Streptomyces sp. isolated from a marine sponge Theonella sp. Tetrahedron Lett. 2016, 57, 4863–4865. [Google Scholar] [CrossRef]
- Chen, H.; Cai, K.; Yao, R. A new macrolactam derivative from the marine actinomycete HF-11225. J. Antibiot. 2018, 71, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.; Li, J.Q.; Zhang, H.J.; Ding, W.J.; Ma, Z.J. Cyclizidine-Type Alkaloids from Streptomyces sp. HNA39. J. Nat. Prod. 2018, 81, 394–399. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Li, J.M.; Qi, H.; Zhang, H.; Zhang, J.; Xiang, W.S.; Wang, J.D.; Wang, X.J. Two new lankacidin-related metabolites from Streptomyces sp. HS-NF-1178. J. Antibiot. 2018, 71, 397–401. [Google Scholar] [CrossRef]
- Zhao, X.L.; Wang, H.; Xue, Z.L.; Li, J.S.; Qi, H.; Zhang, H.; Zhao, T.; Wang, J.D.; Xiang, W.S. Two new glutarimide antibiotics from Streptomyces sp. HS-NF-780. J. Antibiot. 2019, 72, 241–245. [Google Scholar] [CrossRef]
- Yixizhuoma; Ishikawa, N.; Abdelfattah, M.S.; Ishibashi, M. Elmenols C-H, new angucycline derivatives isolated from a culture of Streptomyces sp. IFM 11490. J. Antibiot. 2017, 70, 601–606. [Google Scholar] [CrossRef] [Green Version]
- Son, S.; Jang, M.; Lee, B.; Hong, Y.S.; Ko, S.K.; Jang, J.H.; Ahn, J.S. Ulleungdin, a Lasso Peptide with Cancer Cell Migration Inhibitory Activity Discovered by the Genome Mining Approach. J. Nat. Prod. 2018, 81, 2205–2211. [Google Scholar] [CrossRef]
- Son, S.; Ko, S.K.; Jang, M.; Lee, J.K.; Kwon, M.C.; Kang, D.H.; Ryoo, I.J.; Lee, J.S.; Hong, Y.S.; Kim, B.Y.; et al. Polyketides and Anthranilic Acid Possessing 6-Deoxy-α-l-talopyranose from a Streptomyces Species. J. Nat. Prod. 2017, 80, 1378–1386. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.X.; Hou, G.X.; Luo, J.; Yang, J.; Yan, Y.; Huang, S.X. New phenoxazinone-related alkaloids from strain Streptomyces sp. KIB-H1318. J. Antibiot. 2018, 71, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Dong, M.; Cao, P.; Ma, Y.T.; Luo, J.; Yan, Y.; Li, R.T.; Huang, S.X. A new actinomycin Z analogue with an additional oxygen bridge between chromophore and β-depsipentapeptide from Streptomyces sp. KIB-H714. Nat. Prod. Res. 2019, 33, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Xie, Z.P.; Yang, Q.; Feng, L.L.; Zhang, L.; Zhang, Y.Z.; Li, X.N.; Pescitelli, G.; Zhang, S.M. Kiamycins B and C, unusual bridged angucyclinones from a marine sediment-derived Streptomyces sp. Tetrahedron Lett. 2018, 59, 2176–2180. [Google Scholar] [CrossRef]
- Kawahara, T.; Izumikawa, M.; Kozone, I.; Hashimoto, J.; Kagaya, N.; Koiwai, H.; Komatsu, M.; Fujie, M.; Sato, N.; Ikeda, H.; et al. Neothioviridamide, a Polythioamide Compound Produced by Heterologous Expression of a Streptomyces sp. Cryptic RiPP Biosynthetic Gene Cluster. J. Nat. Prod. 2018, 81, 264–269. [Google Scholar] [CrossRef]
- Lu, D.D.; Ren, J.W.; Du, Q.Q.; Song, Y.J.; Lin, S.Q.; Li, X.; Li, E.W.; Xie, W.D. p-Terphenyls and actinomycins from a Streptomyces sp. associated with the larva of mud dauber wasp. Nat. Prod. Res. 2021, 35, 1869–1873. [Google Scholar] [CrossRef]
- Song, Y.J.; Zheng, H.B.; Peng, A.H.; Ma, J.H.; Lu, D.D.; Li, X.; Zhang, H.Y.; Xie, W.D. Strepantibins A-C: Hexokinase II Inhibitors from a Mud Dauber Wasp Associated Streptomyces sp. J. Nat. Prod. 2019, 82, 1114–1119. [Google Scholar] [CrossRef]
- Cheng, P.; Xu, K.; Chen, Y.C.; Wang, T.T.; Chen, Y.; Yang, C.L.; Ma, S.Y.; Liang, Y.; Ge, H.M.; Jiao, R.H. Cytotoxic aromatic polyketides from an insect derived Streptomyces sp. NA4286. Tetrahedron Lett. 2019, 60, 1706–1709. [Google Scholar] [CrossRef]
- Lu, C.; Zhao, Y.; Jia, W.-Q.; Zhang, H.; Qi, H.; Xiang, W.-S.; Wang, J.-D.; Wang, X.-J. A new anthracycline-type metabolite from Streptomyces sp. NEAU-L3. J. Antibiot. 2017, 70, 1026–1028. [Google Scholar] [CrossRef]
- Kawahara, T.; Fujiwara, T.; Kagaya, N.; Shin-Ya, K. JBIR-150, a novel 20-membered polyene macrolactam from marine-derived Streptomyces sp. OPMA00071. J. Antibiot. 2018, 71, 390–392. [Google Scholar] [CrossRef]
- Abbas, M.; Elshahawi, S.I.; Wang, X.; Ponomareva, L.V.; Sajid, I.; Shaaban, K.A.; Thorson, J.S. Puromycins B-E, Naturally Occurring Amino-Nucleosides Produced by the Himalayan Isolate Streptomyces sp. PU-14G. J. Nat. Prod. 2018, 81, 2560–2566. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chai, W.; Wang, W.; Song, T.; Lian, X.Y.; Zhang, Z. Cytotoxic Bagremycins from Mangrove-Derived Streptomyces sp. Q22. J. Nat. Prod. 2017, 80, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Hayakawa, Y.; Akimoto, M.; Ishikawa, A.; Izawa, M.; Shin-Ya, K. Curromycin A as a GRP78 downregulator and a new cyclic dipeptide from Streptomyces sp. J. Antibiot. 2016, 69, 187–188. [Google Scholar] [CrossRef] [PubMed]
- Nogawa, T.; Okano, A.; Lim, C.L.; Futamura, Y.; Shimizu, T.; Takahashi, S.; Ibrahim, D.; Osada, H. Opantimycin A, a new metabolite isolated from Streptomyces sp. RK88-1355. J. Antibiot. 2017, 70, 222–225. [Google Scholar] [CrossRef]
- Cheng, C.; Othman, E.M.; Fekete, A.; Krischke, M.; Stopper, H.; Edrada-Ebel, R.A.; Mueller, M.J.; Hentschel, U.; Abdelmohsen, U.R. Strepoxazine A, a new cytotoxic phenoxazin from the marine sponge-derived bacterium Streptomyces sp. SBT345. Tetrahedron Lett. 2016, 57, 4196–4199. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Ma, L.; Zhang, L.; Zhang, W.; Zhu, Y.; Chen, Y.; Zhang, W.; Zhang, C. Functional characterization of the halogenase SpmH and discovery of new deschloro-tryptophan dimers. Org. Biomol. Chem. 2019, 17, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Zhang, Q.; Jiang, X.; Ma, L.; Long, T.; Cheng, Z.; Zhang, C.; Zhu, Y. New piericidin derivatives from the marine-derived Streptomyces sp. SCSIO 40063 with cytotoxic activity. Nat. Prod. Res. 2022, 36, 2458–2464. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Gan, L.S.; Ding, W.J.; Chen, Z.; Ma, Z.J. Cytotoxic gephyromycins from the Streptomyces sp. SS13I. Tetrahedron Lett. 2017, 58, 3747–3750. [Google Scholar] [CrossRef]
- Zhang, D.; Jiang, Y.; Li, J.; Ding, W.; Chen, Z.; Ma, Z. Thioquinomycins A-D, novel naphthothiophenediones from the marine-derived Streptomyces sp. SS17F. Tetrahedron 2018, 74, 6150–6154. [Google Scholar] [CrossRef]
- Xu, C.D.; Zhang, H.J.; Ma, Z.J. Pyrimidine Nucleosides from Streptomyces sp. SSA28. J. Nat. Prod. 2019, 82, 2509–2516. [Google Scholar] [CrossRef]
- Lee, B.; Lee, G.-E.; Hwang, G.J.; Heo, K.T.; Lee, J.K.; Jang, J.-P.; Hwang, B.Y.; Jang, J.-H.; Cho, Y.-Y.; Hong, Y.-S. Rubiflavin G, photorubiflavin G, and photorubiflavin E: Novel pluramycin derivatives from Streptomyces sp. W2061 and their anticancer activity against breast cancer cells. J. Antibiot. 2023, 76, 1–7. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, B.Y.; Qin, Y.; Zhuang, L.; Yang, Y.B.; Zhao, L.X. Jiangchuanmycin, a New Pyrrolizidine Analog from Streptomyces sp. YIM S01863. Chem. Biodivers. 2023, 20, e202201240. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wang, Y.; Yang, Y.; Chen, H. Shellmycin A-D, Novel Bioactive Tetrahydroanthra-γ-Pyrone Antibiotics from Marine Streptomyces sp. Shell-016. Mar. Drugs 2020, 18, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El Sayed, K.A. Natural Products as Antiviral Agents. Stud. Nat. Prod. Chem. 2000, 24, 473–572. [Google Scholar] [CrossRef]
- Patridge, E.; Gareiss, P.; Kinch, M.S.; Hoyer, D. An analysis of FDA-approved drugs: Natural products and their derivatives. Drug Discov. Today 2016, 21, 204–207. [Google Scholar] [CrossRef]
- Farmer, P.B.; Suhadolnik, R.J. Nucleoside antibiotics. Biosynthesis of arabinofuranosyladenine by Streptomyces antibioticus. Biochemistry 1972, 11, 911–916. [Google Scholar] [CrossRef]
- Sayed, A.M.; Alhadrami, H.A.; El-Gendy, A.O.; Shamikh, Y.I.; Belbahri, L.; Hassan, H.M.; Abdelmohsen, U.R.; Rateb, M.E. Microbial Natural Products as Potential Inhibitors of SARS-CoV-2 Main Protease (Mpro). Microorganisms 2020, 8, 970. [Google Scholar] [CrossRef]
- Raveh, A.; Delekta, P.C.; Dobry, C.J.; Peng, W.; Schultz, P.J.; Blakely, P.K.; Tai, A.W.; Matainaho, T.; Irani, D.N.; Sherman, D.H.; et al. Discovery of potent broad spectrum antivirals derived from marine actinobacteria. PLoS ONE 2013, 8, e82318. [Google Scholar] [CrossRef] [Green Version]
- Jakubiec-Krzesniak, K.; Rajnisz-Mateusiak, A.; Guspiel, A.; Ziemska, J.; Solecka, J. Secondary Metabolites of Actinomycetes and their Antibacterial, Antifungal and Antiviral Properties. Pol. J. Microbiol. 2018, 67, 259–272. [Google Scholar] [CrossRef] [Green Version]
- Rawal, K.R.; Bariwal, J.; Singh, V. Chemistry and Bioactivities of Aristeromycins: An Overview. Curr. Top. Med. Chem. 2016, 16, 3258–3273. [Google Scholar] [CrossRef]
- Selim, M.S.M.; Abdelhamid, S.A.; Mohamed, S.S. Secondary metabolites and biodiversity of actinomycetes. J. Genet. Eng. Biotechnol. 2021, 19, 72. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef] [PubMed]
- Caly, L.; Druce, J.D.; Catton, M.G.; Jans, D.A.; Wagstaff, K.M. The FDA-approved drug ivermectin inhibits the replication of SARS-CoV-2 in vitro. Antiviral Res. 2020, 178, 104787. [Google Scholar] [CrossRef] [PubMed]
- Euanorasetr, J.; Intra, B.; Thunmrongsiri, N.; Limthongkul, J.; Ubol, S.; Anuegoonpipat, A.; Kurosu, T.; Ikuta, K.; Nihira, T.; Panbangred, W. In vitro antiviral activity of spirotetronate compounds against dengue virus serotype 2. J. Gen. Appl. Microbiol. 2019, 65, 197–203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, L.; Oueis, E.; Kaur, A.; Safaei, N.; Kirsch, S.H.; Gunesch, A.P.; Haid, S.; Rand, U.; Čičin-Šain, L.; Fu, C. Persicamidines—Unprecedented Sesquarterpenoids with Potent Antiviral Bioactivity against Coronaviruses. Angew. Chem. Int. Ed. 2023, 62, e202214595. [Google Scholar] [CrossRef]
- Shuai, H.; Myronovskyi, M.; Nadmid, S.; Luzhetskyy, A. Identification of a Biosynthetic Gene Cluster Responsible for the Production of a New Pyrrolopyrimidine Natural Product-Huimycin. Biomolecules 2020, 10, 1074. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, W.; Wang, K.; Zhang, Z.; Wu, Z.; Shi, L.; Liu, F.; Wan, Z.; Liu, M. Napyradiomycin A4 and Its Relate Compounds, a New Anti-PRV Agent and Their Antibacterial Activities, from Streptomyces kebangsaanensis WS-68302. Molecules 2023, 28, 640. [Google Scholar] [CrossRef]
- Liu, M.; Ren, M.; Zhang, Y.; Wan, Z.; Wang, Y.; Wu, Z.; Wang, K.; Fang, W.; Yang, X. Antiviral activity of benzoheterocyclic compounds from soil-derived Streptomyces jiujiangensis NBERC-24992. Molecules 2023, 28, 878. [Google Scholar] [CrossRef]
- Hao, X.; Li, S.; Wang, G.; Li, J.; Peng, Z.; Zhang, Y.; Yu, L.; Gan, M. Zelkovamycins F and G, Cyclopeptides with Cα-Methyl-threonine Residues, from an Endophytic Kitasatospora sp. J. Nat. Prod. 2022, 85, 1715–1722. [Google Scholar] [CrossRef]
- Kimura, T.; Suga, T.; Kameoka, M.; Ueno, M.; Inahashi, Y.; Matsuo, H.; Iwatsuki, M.; Shigemura, K.; Shiomi, K.; Takahashi, Y.; et al. New tetrahydroquinoline and indoline compounds containing a hydroxy cyclopentenone, virantmycin B and C, produced by Streptomyces sp. AM-2504. J. Antibiot. 2019, 72, 169–173. [Google Scholar] [CrossRef]
- Kim, S.H.; Ha, T.K.Q.; Oh, W.K.; Shin, J.; Oh, D.C. Antiviral Indolosesquiterpenoid Xiamycins C-E from a Halophilic Actinomycete. J. Nat. Prod. 2016, 79, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, J.; Tang, Y.; Guo, Z.; Bai, J.; Wu, L.; Su, J.; Cen, S.; Yu, L.; Zhang, D. Geninthiocins E and F, two new cyclic thiopeptides with antiviral activities from soil-derived Streptomyces sp. CPCC 200267 using OSMAC strategy. J. Antibiot. 2023, 76, 101–104. [Google Scholar] [CrossRef]
- Li, F.; Lu, S.; Xie, X.; Fan, S.; Chen, D.; Wu, S.; He, J. Antiviral properties of extracts of Streptomyces sp. SMU 03 isolated from the feces of Elephas maximus. Fitoterapia 2020, 143, 104600. [Google Scholar] [CrossRef] [PubMed]
- Saito, S.; Funayama, K.; Kato, W.; Okuda, M.; Kawamoto, M.; Matsubara, T.; Sato, T.; Sato, A.; Otsuguro, S.; Sasaki, M. Dihydromaniwamycin E, a Heat-Shock Metabolite from Thermotolerant Streptomyces sp. JA74, Exhibiting Antiviral Activity against Influenza and SARS-CoV-2 Viruses. J. Nat. Prod. 2022, 85, 2583–2591. [Google Scholar] [CrossRef] [PubMed]
- Igarashi, Y.; Yu, L.; Ikeda, M.; Oikawa, T.; Kitani, S.; Nihira, T.; Bayanmunkh, B.; Panbangred, W. Jomthonic Acid A, a Modified Amino Acid from a Soil-Derived Streptomyces. J. Nat. Prod. 2012, 75, 986–990. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Oikawa, T.; Kitani, S.; Nihira, T.; Bayanmunkh, B.; Panbangred, W.; Igarashi, Y. Jomthonic acids B and C, two new modified amino acids from Streptomyces sp. J. Antibiot. 2014, 67, 345–347. [Google Scholar] [CrossRef] [Green Version]
- Inahashi, Y.; Iwatsuki, M.; Ishiyama, A.; Matsumoto, A.; Hirose, T.; Oshita, J.; Sunazuka, T.; Panbangred, W.; Takahashi, Y.; Kaiser, M.; et al. Actinoallolides A–E, New Anti-trypanosomal Macrolides, Produced by an Endophytic Actinomycete, Actinoallomurus fulvus MK10-036. Org. Lett. 2015, 17, 864–867. [Google Scholar] [CrossRef]
- Akiyama, H.; Oku, N.; Harunari, E.; Panbangred, W.; Igarashi, Y. Complete NMR assignment and absolute configuration of k4610422, a norditerpenoid inhibitor of testosterone-5α-reductase originally from Streptosporangium: Rediscovery from a thermophilic Actinomadura. J. Antibiot. 2020, 73, 60–65. [Google Scholar] [CrossRef]
- Lyddiard, D.; Jones, G.L.; Greatrex, B.W. Keeping it simple: Lessons from the golden era of antibiotic discovery. FEMS Microbiol. Lett. 2016, 363, 84. [Google Scholar] [CrossRef] [Green Version]
- Majumder, M.A.A.; Rahman, S.; Cohall, D.; Bharatha, A.; Singh, K.; Haque, M.; Gittens-St Hilaire, M. Antimicrobial Stewardship: Fighting Antimicrobial Resistance and Protecting Global Public Health. Infect. Drug Resist. 2020, 13, 4713. [Google Scholar] [CrossRef]
- Baltz, R.H. Renaissance in antibacterial discovery from actinomycetes. Curr. Opin. Pharm. 2008, 8, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Lazzarini, A.; Cavaletti, L.; Toppo, G.; Marinelli, F. Rare genera of actinomycetes as potential producers of new antibiotics. Antonie Van Leeuwenhoek 2000, 78, 399–405. [Google Scholar] [CrossRef] [PubMed]
- Kohli, I.; Joshi, N.C.; Mohapatra, S.; Varma, A. Extremophile—An Adaptive Strategy for Extreme Conditions and Applications. Curr. Genom. 2020, 21, 96–110. [Google Scholar] [CrossRef] [PubMed]
- Subramani, R.; Aalbersberg, W. Culturable rare Actinomycetes: Diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 2013, 97, 9291–9321. [Google Scholar] [CrossRef]
- Mahajan, G.B.; Balachandran, L. Sources of antibiotics: Hot springs. Biochem. Pharmacol. 2017, 134, 35–41. [Google Scholar] [CrossRef]
- Thawai, C.; Thamsathit, W.; Kudo, T. Planosporangium thailandense sp. nov., isolated from soil from a Thai hot spring. Int. J. Syst. Evol. Microbiol. 2013, 63, 1051–1055. [Google Scholar] [CrossRef] [Green Version]
- Bull, A.T.; Idris, H.; Sanderson, R.; Asenjo, J.; Andrews, B.; Goodfellow, M. High altitude, hyper-arid soils of the Central-Andes harbor mega-diverse communities of actinobacteria. Extremophiles 2017, 22, 47–57. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.; Shi, Y.-L.; Wang, H.; Zhang, T.; Yu, L.-Y.; Sun, H.; Zhang, Y.-Q. Diversity of Bacteria and the Characteristics of Actinobacteria Community Structure in Badain Jaran Desert and Tengger Desert of China. Front. Microbiol. 2018, 9, 1068. [Google Scholar] [CrossRef] [Green Version]
- Li, W.J.; Zhang, Y.G.; Zhang, Y.Q.; Tang, S.K.; Xu, P.; Xu, L.H.; Jiang, C.L. Streptomyces sodiiphilus sp. nov., a novel alkaliphilic actinomycete. Int. J. Syst. Evol. Microbiol. 2005, 55, 1329–1333. [Google Scholar] [CrossRef] [Green Version]
- Hui, M.L.-Y.; Tan, L.T.H.; Letchumanan, V.; He, Y.-W.; Fang, C.-M.; Chan, K.-G.; Law, J.W.F.; Lee, L.H. The Extremophilic Actinobacteria: From Microbes to Medicine. Antibiotics 2021, 10, 682. [Google Scholar] [CrossRef]
- Singh, R.; Dubey, A.K. Diversity and Applications of Endophytic Actinobacteria of Plants in Special and Other Ecological Niches. Front. Microbiol. 2018, 9, 1767. [Google Scholar] [CrossRef] [PubMed]
- Bentley, S.D.; Chater, K.F.; Cerdeno-Tarraga, A.M.; Chalis, G.L.; Thomson, N.R.; James, K.D.; Harris, D.E.; Quail, M.A.; Kieser, H.; Harper, D.; et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 2002, 417, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Hoskisson, P.A.; van Wezel, G.P. Streptomyces coelicolor. Trends Microbiol. 2019, 27, 468–469. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; Van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Weber, T.; Rausch, C.; Lopez, P.; Hoof, I.; Gaykova, V.; Huson, D.H.; Wohlleben, W. CLUSEAN: A computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 2009, 140, 13–17. [Google Scholar] [CrossRef]
- Skinnider, M.A.; Johnston, C.W.; Gunabalasingam, M.; Merwin, N.J.; Kieliszek, A.M.; MacLellan, R.J.; Li, H.; Ranieri, M.R.M.; Webster, A.L.H.; Cao, M.P.T.; et al. Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nat. Comm. 2020, 11, 6058. [Google Scholar] [CrossRef]
- Li, M.H.T.; Ung, P.M.U.; Zajkowski, J.; Garneau-Tsodikova, S.; Sherman, D.H. Automated genome mining for natural products. BMC Bioinform. 2009, 10, 185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starcevic, A.; Zucko, J.; Simunkovic, J.; Long, P.F.; Cullum, J.; Hranueli, D. ClustScan: An integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008, 36, 6882–6892. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Jiang, W.; Lu, Y. New strategies and approaches for engineering biosynthetic gene clusters of microbial natural products. Biotechnol. Adv. 2017, 35, 936–949. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhao, Y.; Huang, C.; Luo, Y. Recent Advances in Silent Gene Cluster Activation in Streptomyces. Front. Bioeng. Biotechnol. 2021, 9, 88. [Google Scholar] [CrossRef]
- Chou, W.K.W.; Fanizza, I.; Uchiyama, T.; Komatsu, M.; Ikeda, H.; Cane, D.E. Genome mining in Streptomyces avermitilis: Cloning and characterization of SAV_76, the synthase for a new sesquiterpene, avermitilol. J. Am. Chem. Soc. 2010, 132, 8850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring structural diversity of microbe secondary metabolites using OSMAC strategy: A literature review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef] [Green Version]
- Romano, S.; Jackson, S.A.; Patry, S.; Dobson, A.D.W. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asamizu, S.; Ozaki, T.; Teramoto, K.; Satoh, K.; Onaka, H. Killing of Mycolic Acid-Containing Bacteria Aborted Induction of Antibiotic Production by Streptomyces in Combined-Culture. PLoS ONE 2015, 10, e0142372. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Li, X.; Li, Z.; Zhan, X.; Mao, X.; Li, Y. The Application of Regulatory Cascades in Streptomyces: Yield Enhancement and Metabolite Mining. Front. Microbiol. 2020, 11, 406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tawfike, A.; Attia, E.Z.; Desoukey, S.Y.; Hajjar, D.; Makki, A.A.; Schupp, P.J.; Edrada-Ebel, R.A.; Abdelmohsen, U.R. New bioactive metabolites from the elicited marine sponge-derived bacterium Actinokineospora spheciospongiae sp. nov. AMB Express 2019, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Li, Y.; Banakar, S.P.; Liu, L.; Shao, C.; Li, Z.; Wang, C. New metabolites from the Co-culture of marine-derived actinomycete Streptomyces rochei MB037 and fungus Rhinocladiella similis 35. Front. Microbiol. 2019, 10, 915. [Google Scholar] [CrossRef] [Green Version]
- Tyurin, A.P.; Alferova, V.A.; Korshun, V.A. Chemical Elicitors of Antibiotic Biosynthesis in Actinomycetes. Microorganisms 2018, 6, 52. [Google Scholar] [CrossRef] [Green Version]
- Zong, G.; Fu, J.; Zhang, P.; Zhang, W.; Xu, Y.; Cao, G.; Zhang, R. Use of elicitors to enhance or activate the antibiotic production in Streptomyces. Crit. Rev. Biotechnol. 2022, 42, 1260–1283. [Google Scholar] [CrossRef]
- Choi, S.U.; Lee, C.K.; Hwang, Y.I.; Kinosita, H.; Nihira, T. γ-Butyrolactone autoregulators and receptor proteins in non-Streptomyces actinomycetes producing commercially important secondary metabolites. Arch. Microbiol. 2003, 180, 303–307. [Google Scholar] [CrossRef]
- Onaka, H. Novel antibiotic screening methods to awaken silent or cryptic secondary metabolic pathways in actinomycetes. J. Ant. 2017, 70, 865–870. [Google Scholar] [CrossRef] [PubMed]
- Dashti, Y.; Grkovic, T.; Abdelmohsen, U.R.; Hentschel, U.; Quinn, R.J. Actinomycete Metabolome Induction/Suppression with N-Acetylglucosamine. J. Nat. Prod. 2017, 80, 828–836. [Google Scholar] [CrossRef] [PubMed]
- Mast, Y.; Stegmann, E. Actinomycetes: The Antibiotics Producers. Antibiotics 2019, 8, 105. [Google Scholar] [CrossRef] [Green Version]
- Jose, P.A.; Jha, B. New Dimensions of Research on Actinomycetes: Quest for Next Generation Antibiotics. Front. Microbiol. 2016, 7, 1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gadelhak, G.G.; El-Tarabily, K.A.; Al-Kaabi, F.K. Insect Control Using Chitinolytic Soil Actinomycetes as Bio control Agents. Int. J. Agri. Biol. 2005, 7, 627–633. [Google Scholar]
- Lam, K.S. Discovery of novel metabolites from marine actinomycetes. Curr. Opin. Microbiol. 2006, 9, 245–251. [Google Scholar] [CrossRef]
- Vijayabharathi, R.; Kumari, B.R.; Sathya, A.; Srinivas, V.; Abhishek, R.; Sharma, H.; Gopalakrishnan, S. Biological activity of entomopathogenic actinomycetes against lepidopteran insects (Noctuidae: Lepidoptera). Can. J. Plant Sci. 2014, 94, 759–769. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.-S.; Kim, H.-J.; Lee, H.-S.; Kim, P.; Kim, E.-S. Genome mining of rare actinomycetes and cryptic pathway awakening. Process. Biochem. 2015, 50, 1184–1193. [Google Scholar] [CrossRef]
- Peng, Q.; Gao, G.; Lü, J.; Long, Q.; Chen, X.; Zhang, F.; Xu, M.; Liu, K.; Wang, Y.; Deng, Z.; et al. Engineered Streptomyces lividans Strains for Optimal Identification and Expression of Cryptic Biosynthetic Gene Clusters. Front. Microbiol. 2018, 9, 3042. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B.K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef]
- Gomez-Escribano, J.P.; Bibb, M.J. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Microb. Biotechnol. 2011, 4, 207–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, D.; Okada, B.K.; Wu, Y.; Xu, F.; Seyedsayamdost, M.R. Recent advances in activating silent biosynthetic gene clusters in bacteria. Curr. Opin. Microbiol. 2018, 45, 156–163. [Google Scholar] [CrossRef] [PubMed]
- Salcedo, R.G.; Olano, C.; Gómez, C.; Fernández, R.; Braña, A.F.; Méndez, C.; Calle, F.; Salas, J.A. Characterization and engineering of the biosynthesis gene cluster for antitumor macrolides PM100117 and PM100118 from a marine actinobacteria: Generation of a novel improved derivative. Microb. Cell Factories 2016, 15, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoshino, S.; Onaka, H.; Abe, I. Activation of silent biosynthetic pathways and discovery of novel secondary metabolites in actinomycetes by co-culture with mycolic acid-containing bacteria. J. Ind. Microbiol. Biotechnol. 2018, 46, 363–374. [Google Scholar] [CrossRef] [PubMed]




| Organism (s) | Compound Name (s) | Reference (s) |
|---|---|---|
| Actinoallomurus sp. ID145113, 145206, 145754 | Paramagnetoquinones A–C | [66] |
| Actinomadura atramentaria NBRC 14695 | Cinnamycin B | [67] |
| Actinomadura sp. KC191 | Actinomadurol | [68] |
| Amycolatopsis sp. IRD-009 | Pradimicin-IRD | [69] |
| Amycolatopsis sp. M39 | Macrotermycins A & C | [70] |
| Amycolatopsis sp. MCC0218 | Enceleamycins A–C | [71] |
| Amycolatopsis sp. ML1-hF4 | Pargamicins B–D | [72] |
| Kibdelosporangium phytohabitans XY-R10 | Maipomycin A (MaiA) | [73] |
| Kitasatospora sp. MG372-hF19 | Biospolides C-E | [74] |
| Kocuria marina CMG S2 | Kocumarin | [75] |
| Micromonospora carbonacea LS276 | Tetrocarcin Q | [76,77] |
| Micromonospora harpali SCSIO GJ089 | Icrosporanate A, Tetrocarcin P, Microsporanates B–F | [78] |
| Micromonospora chalcea FIM 02 | Rakicidins G–I | [79] |
| Micromonospora sp. UR56 + Actinokineospora sp. EG49 | Phenazine-1-carboxylic acid, Aestivophoenin C, Methyl saphenate | [80] |
| Micromonospora sp. WMMB-235 + Rhodococcus sp. WMMA-185 | Keyicin | [81] |
| Micromonospora sp. 5-297 | Tetrocarcins N | [82] |
| Micromonospora sp. CA-214671 | Phocoenamicin B | [83] |
| Micromonospora sp. RJA4480 | Sporalactam B | [84] |
| Micromonospora sp. SCSIO 07395 | Microechmycin A | [85] |
| Micromonospora sp. TP-A0468 | 16-demethylrifamycin S | [86] |
| Micromonospora endolithica | loseolamycins A1, A2 | [87] |
| Micromonospora yangpuensis DSM 45577 | Yangpumicins F, G | [88] |
| Nocardiopsis sp. LX-1 | Nocarpyrroline A, Isoflavonoid E | [89] |
| Nocardiopsis sp. SCA30 | 1-acetyl-4-4(hydroxyphenyl)piperazine | [90] |
| Pseudonocardia carboxydivorans M227 | Branimycins B, C | [91] |
| Streptomonospora sp. PA3 | Persiamycin A | [92] |
| Streptomyces aculeolatus PTM-029 | Napyradiomycins (3,7,9,10,12) | [93,94] |
| Streptomyces aculeolatus PTM-420 | Napyradiomycins (1,2,4–6,8,11) | [93] |
| Streptomyces albus 4N24 | Benzanthric acid | [95] |
| Streptomyces albus MAB56 | 12-methyltetradecanoic acid, Palmitic acid, Tridecanoic acid | [96] |
| Streptomyces armeniacus DSM 43125 | Armeniaspirol analogues 1–6, 9–12 | [94] |
| Streptomyces bacillaris MBTC38 | Lactoquinomycin A | [97] |
| Streptomyces californicus ADR1 | Methanoazulen-9-ol, decahydro-2, 2, 4, 8-tetramethyl-stereoisomer (Sesquiterpene) | [98] |
| Streptomyces coelicolor LY001 | 3-(3,5-dichloro-4-hydroxyphenyl) propanoic acid, 3-(3,5-dichloro-4-hydroxyphenyl) propanoic acid methyl ester, 3-(3-chloro-4-hydroxyphenyl) propanoic acid | [99] |
| Streptomyces diacarni LHW51701 | Chlocarbazomycins (CCBs) A–D | [100] |
| Streptomyces fradiae MM456M-mF7 | Fradiamines B | [101] |
| Streptomyces globisporus sp. WA5-2-37 | Actinomycin X2, Collismycin A | [102] |
| Streptomyces globisporus subsp. globisporus | Globimycin | [103] |
| Streptomyces griseoviridis PU-KB10–4 | Mitomycin C | [104] |
| Streptomyces hyaluromycini MB-PO13 | Rubromycins CA1, CA2 | [105] |
| Streptomyces koyangensis SCSIO 5802 | Neoabyssomicins F, G | [106] |
| Streptomyces malachitospinus ITD-35 | 3-octanone, neopentyl, Isothiocyanate, 2-methyl butyl isothiocyanate | [13] |
| Streptomyces microflavus (MBTI36) | Chromomycin A9, Ap, A2, A3 | [107] |
| Streptomyces lunaelactis MM109T | Lunaemycins A, B1, D | [108] |
| Streptomyces lusitanus OUCT16-27 | Grincamycin L | [109] |
| Streptomyces palmae CMU-AB204T | Phenyl alkenoic acids C, D Phenyl alkenoic acids E, F | [110] |
| Streptomyces qinglanensis 172205 | 15R-17,18-dehydroxantholipin | [111] |
| Streptomyces zhaozhouensis 208DD-064 | Streptopyrroles B, C | [112] |
| Streptomyces sp. 120454 | Mayamycin B | [113] |
| Streptomyces sp. 182SMLY | N-acetyl-N-demethylmayamycin | [114] |
| Streptomyces sp. 7NS3 | Emycin A | [115] |
| Streptomyces sp. 8P21H-1 | Desulphurzing, Griseoviridin, Griseoviridin | [116] |
| Streptomyces sp. AD-3-6 | Nybomycins D | [117] |
| Streptomyces sp. ADI91-18, ADI95-16 | Linearmycins | [117] |
| Streptomyces sp. ADI97-07 | Clavams | [117] |
| Streptomyces sp. ADI98-12 | Griselimycin, Kirromycin | [117] |
| Streptomyces sp. B9173 | Flaviogeranin B1, B, Flaviogeranin D, Flaviogeranin C2 | [118] |
| Streptomyces sp. CA-271078 | Napyradiomycins 2, D1 | [119] |
| Streptomyces sp. CPCC 204980 | Cervinomycins B1–B4 | [120] |
| Streptomyces sp. DSM14386 | Svetamycins C, G | [121] |
| Streptomyces sp. FJS31-2 | Zunyimycins B, C | [122] |
| Streptomyces sp. GKU 220 | Rakicidin F | [123] |
| Stretomyces sp. HCCB11876 | Quinomycins I, J | [124] |
| Streptomyces sp. HK-2006-1 | Aldgamycin O | [125] |
| Streptomyces sp. HN-A124 | Cysrabelomycin | [126] |
| Streptomyces sp. Hu103 | Anulamycins A-D | [127] |
| Streptomyces sp. HZP-2216E | 23-O-butyrylbafilomycin D | [128] |
| Streptomyces sp. IB201691-2A | Baikalomycins A–C | [129] |
| Streptomyces sp. ICN19 | Ala-geninthiocin | [130] |
| Streptomyces sp. KCB13F003 | Ulleungmycin A, B | [131] |
| Streptomyces sp. KCB14A132 | Enamidonins B, C | [132] |
| Streptomyces sp. MM168-141F8 | Quadoctomycin | [133] |
| Streptomyces sp. MNP32 | Phenol, 3,5-bis(1,1-dimethylethyl)-1,1′-Biphenyl]-2,3′-diol, 3,4′,5,6′-tetrakis(1,1-dimethylethyl) | [134] |
| Streptomyces sp. MUSC 125 | Thiophene, 2-butyl-5-ethyl, 1-Heptyn-3-ol, 8-[N-Aziridylethylamino]-2-6,dimethyloctene-2, Pyrrolo[1,2-a]pyrazine-1,4-dion,hexahydro, and 2,4-Dihydroxy-6-propylbenzoic acid | [42] |
| Streptomyces sp. NA06554 | Borrelidin J | [135] |
| Streptomyces sp. NA07423 | Nagimycins A, B | [136] |
| Streptomyces sp. OPOK_MB_B11 | Azalomycin F4a 2-ethylpentyl ester | [137] |
| Streptomyces sp. PBR11 | 1-Tetradecanol phenol, 2,5-bis(1,1-dimethylethyl n-pentadecanol, 1-nonadecene, and pyrrolo[1,2-a]pyrazine-1,4-dione hexahydro-3-(phenylmethyl). | [138] |
| Streptomyces sp. PNM-9 | 2-methyl-N-(2′-phenylethyl)-butanamide 3-methyl-N-(2′-phenylethyl)-butanamide | [139] |
| Streptomyces sp. SBT345 | Ageloline A | [140] |
| Streptomyces sp. SCSIO 41399 | Isotirandamycin B | [141] |
| Streptomyces sp. sima1_6 | Echinomycin | [137] |
| Streptomyces sp. SN0280 | Streptoone A | [142] |
| Streptomyces sp. SM01 | Picolinamycin | [143] |
| Streptomyces sp. SS | Sansanmycin Q | [144] |
| Streptomyces sp. Stup16_B49.2 | Echinomycin | [137] |
| Streptomyces sp. Stup16_B146 | TPU-0037-C | [137] |
| Streptomyces sp. Stup18_J70 | Bafilomycin A1 derivative | [137] |
| Streptomyces sp. UICC B-92 | 4-O-glucosyl,1-carboxyl-phenazine | [145] |
| Streptomyces sp. W367A | W367A | [137] |
| Streptomyces sp. XMA39 | Strepoxepinmycins A–D | [146] |
| Streptomyces sp. YINM00001 | Peperodione, Peperophthalene | [147] |
| Streptomyces sp. ZZ1118 | Streptoindoles A–D | [148] |
| Streptomyces sp. ZZ741 | Streptoglutarimides A–J | [149] |
| Streptomycete clade MAR4 CNY-960, CNS-284 | Marinocyanins A–F | [150] |
| Streptosporangium sp. SANK 60,501 | Muraminomicins A, B, C, D, E1, E2, F | [151] |
| Thermoactinomyces vulgaris ISCAR 2354 | Thermoactinoamide A | [152] |
| Verrucosispora sp. FIM06-0036 | 2-ethylhexyl 1H-imidazole-4-carboxylate | [100] |
| Verrucosispora sp. SCSIO 07399 | Kendomycins B–D | [153] |
| Organism (s) | Compound Name (s) | Reference (s) |
|---|---|---|
| Actinokineospora bangkokensis 44EHWT | Thailandins A & B | [159] |
| Actinomadura sp. BCC 35,430 | Actinomadurone | [161] |
| Amycolatopsis sp. M39 | Macrotermycins A & D | [70] |
| Acrocarpospora punica 04107M | Acrocarposporins A, B, D, and E | [162] |
| Dermabacter vaginalis AD1-86 | Dermazolium A | [163] |
| Micromonospora sp. UR56 + Actinokineospora sp. EG49 | Phenazine-1-carboxylic acid, Aestivophoenin c, Methyl saphenate | [80] |
| Nocardia abscessus IFM 10029 | Nabscessins A, B | [164] |
| Nocardiopsis sp. LX-1 | Isoflavonoid E | [89] |
| Pseudonocardia autotrophica KCTC9441 | Polyene B1 | [165] |
| Saccharothrix yanglingensis Hhs.015 | 10-deoxyfungichromin (WH02) | [166] |
| Streptomyces albidoflavus STV1572a | 1-heneicosanol | [167] |
| Streptomyces albolongus YIM 101047 | 19-methoxybafilomycin C1 amide | [168] |
| Streptomyces antibioticus strain 200-09 | Kitamycin C | [169] |
| Streptomyces caniferus GUA-06-05-006A | PM100117, PM100118 | [170] |
| Streptomyces griseoviridis PU-KB10–4 | Mitomycin C | [104] |
| Streptomyces hyaluromycini MB-PO13 | Rubromycins CA1, CA2 | [105] |
| Streptomyces morookaense AM25 | Gloeosporiocide | [171] |
| Streptomyces sp. 1H-XA2 | Furamycins I, II | [172] |
| Streptomyces sp. ADI91-18 | Linearmycins | [117] |
| Streptomyces sp. ADI95-16 | Linearmycins | [117] |
| Streptomyces sp. ADI96-02 | Cycloheximide, Galbonolides | [117] |
| Streptomyces sp. ADI96-15 | Candicidin | [117] |
| Streptomyces sp. ADI97-07 | Galbonolides | [117] |
| Streptomyces sp. ADI98-12 | Candicidin | [117] |
| Streptomyces sp. BV410 | Staurosporine | [173] |
| Streptomyces sp. CB09030 | LOB A, B, H8 | [174] |
| Streptomyces sp. CT37 | Legonimide 1, 1H-indole-3-carbaldehyde | [175] |
| Streptomyces sp. FX13 | Oligomycin A | [176] |
| Streptomyces sp. HAAG3-15 | Azalomycin B | [177] |
| Streptomyces sp. KIB-H869 | Hygrolidin-type macrolide | [178] |
| Streptomyces sp. MUSC 125 | Thiophene, 2-butyl-5-ethyl, 1-Heptyn-3-ol, Pyrrolo[1,2-a] pyrazine-1,4-dion,hexahydro, 9,9-Dimethyl-3, 7-diazabicyclo[3.3.1]nonane, 2,4-Dihydroxy-6-propylbenzoic acid | [42] |
| Streptomyces sp. PBR11 | 1-tetradecanol, n-pentadecanol, 1-nonadecene, pyrrolo [1,2-a]pyrazine-1,4-dione hexahydro-3-(phenylmethyl) | [138] |
| Streptomyces sp. QHH-9511 | 6-deoxy-13-hydroxy-8,11-dione Dihydrogranaticin A Granaticin A, B | [179] |
| Streptomyces sp. SN0280 | Streptoone B | [142] |
| Streptomyces sp. SY1965 | Streptothiazomycin A, Streptodiketopiperazines A, B, [2-hydroxy-1-(hydroxymethyl)ethyl]-2-methoxybenzamide, salicylamide, 4-hydroxymethyl benzoate, Spoxazomicin C | [180] |
| Streptomyces sp. TR1341 | Filipin, Fungichromin, Actinomycin X2 | [181] |
| Streptomyces sp. TT3 | Actinorhodin | [182] |
| Streptomyces sp. YO15-A001 | YO-001A | [183] |
| Streptomyces albus CAI-21 | Organophosphate | [184] |
| Streptomyces coelicolor LY001 | diketopiperazine alkaloids cyclo (l-Phe-trans-4-OH-l-Pro), cyclo(l-Phe-cis-4-OH-d-Pro) | [99] |
| Streptomyces palmae CMU-AB204T | Phenyl alkenoic acids A,B, Anguinomycin A, Leptomycin A, Actinopyrone A | [110] |
| Streptomyces qinglanensis 172205 | 15R-17,18-dehydroxantholipin | [111] |
| Streptomycete clade MAR4 CNY-960 and CNS-284 | Marinocyanins A | [150] |
| Umezawaea sp. RD066910 + Tsukamurella pulmonis TP-B0596 | Umezawamides A | [185] |
| Organism (s) | Compound Name (s) | Reference (s) |
|---|---|---|
| Actinoalloteichus hymeniacidonis 179DD-027 | Dokdolipid B | [198] |
| Actinomadura sp. K13-0306 | Sagamilactam | [199] |
| Actinomadura sp. K4S16 | Nonthmicin, Ecteinamycin | [200] |
| Actinosynnema pretiosum HGF052::asm18 | Actinosynneptide A, B | [201] |
| Amycolatopsis sp. IRD-009 | Pradimicin-IRD | [69] |
| Amycolatopsis alba DSM 44,262 | Thioalbamide | [202] |
| Catenuloplanes sp. RD067331 + Tsukamurella pulmonis TP-B059 | Catenulobactin B | [203] |
| Micromonospora sp. UR56 + Actinokineospora sp. EG49 | Phenazine-1-carboxylic acid, Aestivophoenin c, Methyl saphenate | [80,204] |
| Micromonospora carbonacea LS276 | Tetrocarcin Q | [76] |
| Micromonospora zhangzhouensis HM134T | 7E,11E)-6-hydroxy-1-isopropyl-11-(methoxycarbonyl)-4-methylene-1,2,3,4,4a,5,6,9,10,12a-decahydrobenzo[10]annulene-7-carboxylic acid | [205] |
| Micromonospora matsumotoense M-412 | Paulomycin G | [206] |
| Micromonospora aurantiaca 110B | Isoflavonoid glycosides 1-3 | [207] |
| Micromonospora chalcea FIM 02-523 | Rakicidins G–I | [208] |
| Micromonospora sp. FIM05328 | FW05328-1 | [209] |
| Micromonospora sp. HS-HM-036 | Naphthalenepropanoic acid analog | [210] |
| Micromonospora yangpuensis DSM 45577 | Yangpumicin A | [211] |
| Micromonospora yangpuensis DSM 45577 | Yangpumicins F, G | [88] |
| Nocardiopsis alba KM6-1 | Isomethoxyneihumicin | [212] |
| Nocardiopsis sp. CG3 | Kenalactams A, E | [213] |
| Nonomuraea endophytica GW58/450 | Karamomycins B, C | [214] |
| Nonomuraea sp. AKA32 | Akazamicin | [215] |
| Saccharomonospora sp. UR22 + Dietzia sp. UR66 | Saccharomonosporine A | [216] |
| Streptomonospora sp. PA3 | Persiamycin A | [92] |
| Streptomyces albolongus YIM 101047 | 19-methoxybafilomycin C1 amide, 21-deoxybafilomycin A1 | [168] |
| Streptomyces ardesiacus 156VN-095 | Urdamycins W, X, Grincamycin U | [217] |
| Streptomyces bacillaris | 2,4-di-tert-butylphenol | [218] |
| Streptomycete clade MAR4 CNY-960 and CNS-284 | Marinocyanins A–F | [150] |
| Streptomyces qinglanensis 172205 | 15R-17,18-dehydroxantholipin, (3E,5E,7E)-3-methyldeca-3,5,7-triene-2,9-dione, Qinlactone A–B | [111] |
| Streptomyces cacaoi subsp. asoensis H2S5 | Trienomycins J–L | [219] |
| Streptomyces caniferus GUA-06-05-006A | PM100117, PM100118 | [220] |
| Streptomyces coelicolor M1146 | Staurosporines M1, M2 | [221] |
| Streptomyces curacoi NBRC 12761 | Curacozole | [222] |
| Streptomyces cyaneofuscatus M-157 | 3-Hydroxyquinaldic acid derivative 1 | [223] |
| Streptomyces griseoviridis 2464-S5 | Prodigiosin R2 | [224] |
| Streptomyces seoulensis A01 | Ansaseomycins A, B | [225] |
| Streptomyces subflavus subsp. irumaensis AM-3603 | Bisoxazolomycin A | [226] |
| Streptomyces violascens YIM 100225 | Albaflavenoid | [227] |
| Streptomyces violascens YIM100212 | Fusicomycin A, B, Isofusicomycin A | [228] |
| Streptomyces sp. 166 | Sekgranaticin | [229] |
| Streptomyces sp. 182SMLY | N-acetyl-N-demethylmayamycin, Streptoanthraquinone A | [230] |
| Streptomyces sp. 1H-GS5 | Spectinabilin derivative 1 | [231] |
| Streptomyces sp. A65 | Streptocarbazoles C, 2040-epi-K252d, 0-epi-K252d | [232] |
| Streptomyces sp. AD-3 | Nybomycins D | [233] |
| Streptomyces sp. ADI92-24 | Tomaymycin | [117] |
| Streptomyces sp. ADI95-17 | Naringenin | [117] |
| Streptomyces sp. ADI96-02 | Echinomycin | [117] |
| Streptomyces sp. ADI96-15 | Antimycin, Lobophorins | [117] |
| Streptomyces sp. ADI97-07 | Neocarzilin, Antimycin | [117] |
| Streptomyces sp. ADI98-10 | Actinomycin | [117] |
| Streptomyces sp. B9173 | Flaviogeranin B1, B, C2, D | [118] |
| Streptomyces sp. BB47 | Antarlides F–H | [234] |
| Streptomyces sp. CHQ-64 ΔrdmF | Geranylpyrrol A, Piericidin F | [235] |
| Streptomyces sp. CMAA 1527 | Cinerubin B | [15] |
| Streptomyces sp. CMAA 1653 | Actinomycin V | [15,120] |
| Streptomyces sp. CPCC 204,980 | Cervinomycins B1-B4 | [120] |
| Streptomyces sp. DT-A61 | 9-hydroxy-K252c, 3-hydroxy-K252c, 3-hydroxy-7-methoxy-K252c, Nacetylholyrine A, 3-hydroxyholyrine A, 3′-O-demethyl-4′-N-demethyl-4′-N-acetyl-4′-epi-staurosporine, Streptocarbazoles D, E | [236] |
| Streptomyces sp. EGY1 | Sharkquinone | [237] |
| Streptomyces sp. GIC10-1 | Bafilomycin M | [238] |
| Streptomyces sp. GKU 220 | Rakicidin F | [123] |
| Streptomyces sp. HF-11225 | Nivelactam B | [239] |
| Streptomyces sp. HN-A124 | Cysrabelomycin | [126] |
| Streptomyces sp. HNA39 | Cyclizidines B–I | [240] |
| Streptomyces sp. HS-NF-1178 | 218-seco-lankacidinols A, B | [241] |
| Streptomyces sp. HS-NF-780 | 9-methylstreptimidone 2-α-d-glucopyranoside, ydroxyiso-9-methylstreptimidone | [242] |
| Streptomyces sp. HZP-2216E | 23-O-butyrylbafilomycin D | [81,128] |
| Streptomyces sp. ICN19 | Ala-geninthiocin | [130] |
| Streptomyces sp. IFM 11490 | Elmenols G | [243] |
| Streptomyces sp. KCB13F003 | Ulleungdin | [244] |
| Streptomyces sp. KCB13F030 | Ulleungoside | [245] |
| Streptomyces sp. KIB-H1318 | Phenoxazinone-related alkaloid 2 | [246] |
| Streptomyces sp. KIB-H714 | Actinomycin Z6 | [247] |
| Streptomyces sp. M268 | Kiamycin B | [248] |
| Streptomyces sp. MSB090213SC12 | Neothioviridamide | [249] |
| Streptomyces sp. MUSC 125 | 8-[N-Aziridylethylamino]-2-6,dimethyloctene-2 | [42] |
| Streptomyces sp. N1510.2 | Strepantibin D | [250] |
| Streptomyces sp. N1510.2 | Strepantibins A–C | [251] |
| Streptomyces sp. NA4286 | Murayaquinone D | [252] |
| Streptomyces sp. NEAU-L3 | Tetracenoquinocin A | [253] |
| Streptomyces sp. OPMA00071 | JBIR-150 | [254] |
| Streptomyces sp. PU-14G | Puromycins C, D | [255] |
| Streptomyces sp. PU-KB10-4 | 4-hydroxycinnamide | [104] |
| Streptomyces sp. Q22 | Bagremycins C | [256] |
| Streptomyces sp. RAI364 | Curromycin A | [257] |
| Streptomyces sp. RK88 | Opantimycin A | [258] |
| Streptomyces sp. SBT345 | Strepoxazine A | [259] |
| Streptomyces sp. SCSIO 03032 | Indimicins F, G, Spiroindimicins G, H | [260] |
| Streptomyces sp. SCSIO 1666/17C4 (engineered) | l-rhodinose-l-rhodinose-2-deoxy-l-fucose-10-decarbomethoxy-ε-rhodomycinone | [219] |
| Streptomyces sp. SCSIO 40063 | Piericidins A5, G1 | [261] |
| Streptomyces sp. SCSIO 41399 | Aranciamycin K, Isotirandamycin B | [141] |
| Streptomyces sp. SS13I | Ephyromycins B, C | [262] |
| Streptomyces sp. SS17F | Thioquinomycins A, C, D | [263] |
| Streptomyces sp. SSA28 | Cytosaminomycin E | [264] |
| Streptomyces sp. TR1341 | Filipin, Fungichromin, Actinomycin X2 | [181] |
| Streptomyces sp. XMA39 | Strepoxepinmycins A–D | [146] |
| Streptomyces sp. W2061 | Rubiflavin G Photorubiflavin G, E | [265] |
| Streptomyces sp. YIM S01863 | Jiangchuanmycin | [266] |
| Streptomyces sp. ZZ741 | Streptoglutarimides H | [149] |
| Streptomyces OPOK_MB_A9 | Milbemycin-α8 | [137] |
| Streptomyces sp. shell-016 | Shellmycin A–D | [267] |
| Umezawaea sp. RD066910 + Tsukamurella pulmonis TP-B0596 | Umezawamides A & B | [185] |
| Verrucosispora sp. SCSIO 07399 | Kendomycins B-D | [153] |
| Organism (s) | Compound Name (s) | Reference (s) |
|---|---|---|
| Actinomadura sp. 2EPS | Decatromicins | [278] |
| Kibdelosporangium persicum | Persicamidines A–E | [279] |
| Kutzneria albida DSM 43870 | Huimycin | [106,280] |
| Streptomyces kebangsaanensis WS-68302 | Napyradiomycin A4, A80915 H | [281] |
| Streptomyces jiujiangensis NBERC-24992 | Virantmycins D–G | [282] |
| Streptomyces bacillaris | Zelkovamycins F, G | [283] |
| Streptomyces koyangensis SCSIO 5802 | Neoabyssomicins F, G | [106,284] |
| Streptomyces sp. AM-2504 | Virantmycins B | [284,285] |
| Streptomyces sp. CPCC 200267 | Geninthiocins E, F | [286] |
| Streptomyces sp. HK18 | Xiamycins D | [285,287] |
| Streptomyces sp. JA74 | Dihydromaniwamycin E | [288] |
| Streptomyces sp. SMU 03 | dichloromethane extracts (DCME) | [287] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngamcharungchit, C.; Chaimusik, N.; Panbangred, W.; Euanorasetr, J.; Intra, B. Bioactive Metabolites from Terrestrial and Marine Actinomycetes. Molecules 2023, 28, 5915. https://doi.org/10.3390/molecules28155915
Ngamcharungchit C, Chaimusik N, Panbangred W, Euanorasetr J, Intra B. Bioactive Metabolites from Terrestrial and Marine Actinomycetes. Molecules. 2023; 28(15):5915. https://doi.org/10.3390/molecules28155915
Chicago/Turabian StyleNgamcharungchit, Chananan, Nutsuda Chaimusik, Watanalai Panbangred, Jirayut Euanorasetr, and Bungonsiri Intra. 2023. "Bioactive Metabolites from Terrestrial and Marine Actinomycetes" Molecules 28, no. 15: 5915. https://doi.org/10.3390/molecules28155915